October 15, 2010 (Vol. 30, No. 18)
Emergence of Synthetic Biology Necessitates New Solutions to Generate Molecules
Genome engineering has expanded to cover myriad applications including the analysis of complex pathways, the construction of new biological parts, and the re-design of existing biological systems. All these areas require the precise and concerted assembly of multiple DNA fragments of various sizes, including chromosomes.
Current commercial cloning products are not robust enough to support the assembly of very large or very small genetic elements or a combination of both. In addition, current strategies are not flexible enough to allow further modifications to the original design without having to undergo complicated cloning strategies.
In this article we present a set of technologies from Invitrogen, part of Life Technologies, that allow the seamless, simultaneous, flexible, and highly efficient assembly of genetic material, designed for a wide dynamic size range. The assembly can be performed either in vitro using an enzymatic mix or within the living cells.
In Vivo Recombineering
The in vivo approach requires the transformation into yeast of two or more linear DNA molecules that share unique common end sequences. End-homology can even be provided in trans by overlapping oligonucleotides. If one of the molecules harbors replication and selection elements for yeast and E. coli, then circular episomes up to 110 kilo base-pairs (kbp) in length can be assembled in yeast and transferred to E. coli for downstream applications.
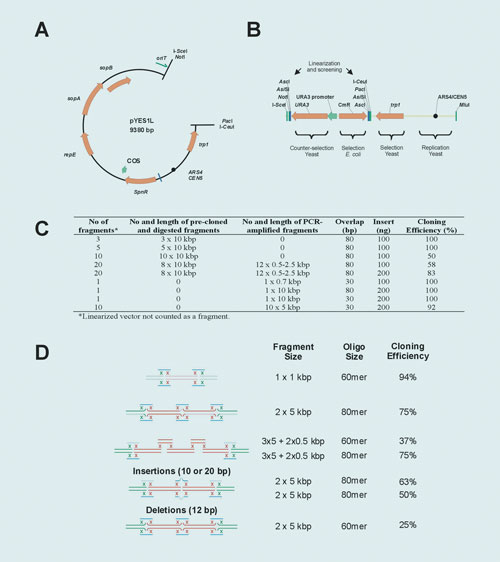
For this purpose a new vector, pYES1L, was constructed (Figure 1A). The plasmid contains a S. cerevisiae’s centromere and the trp1 gene for selection in yeast. In addition F´ ori, the spectinomycin resistance gene, an oriT, and the bacteriophage λ COS site were added for manipulation in E. coli. Rare restriction sites were added for cloning and mapping. We also designed a cassette to convert any E. coli plasmid into a YAC episome, with yeast features similar to pYES1L plus the URA3 gene as an optional counter-selectable marker (Figure 2B).
The system was tested by simultaneously assembling up to 10 linear fragments in a predetermined order into pYES1L (Figure 1C). Adjacent DNA fragments, which shared 30 to 80 base pairs (bp) of end-homology, had been previously cloned into pACYC184 or directly amplified by PCR.
DNA (up to 200 ng) was combined with S. cerevisiae MaV203 competent cells (trp1-901; URA3) and plated onto tryptophan-free CMS agar medium. Clones were verified by colony-PCR and sequencing and/or transferred back to E. coli by directly electroporating colony lysates for restriction profiling. Cloning efficiencies ranged from 58 to 100%.
The approach has also been applied to recombine fragments that do not share end-homology. Here, homology is provided in trans by complementary oligonucleotides that overlap both fragments (Figure 1D). The method is advantageous for re-using fragments in a new sequence context, for cloning DNA targets that cannot be readily amplified by PCR, or for editing the fragment junctions generating end imperfections.
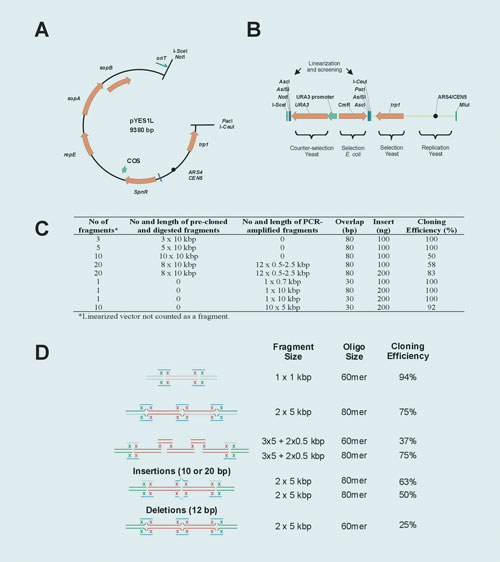
Figure 1. Features and performance of the in vivo recombineering system: (A) Map of pYES1L. SpnR, spectinomycin resistance gene, repE-sopA-sopB genes are required for replication in E. coli; ARS4-CEN5, S. cerevisiae’s chromosome II centromere; trp1, gene encoding yeast’s phosphoribosylanthranilate isomerase; oriT, F’ origin of transference; COS, bacteriophage lambda cos site. (B) YAC conversion cassette. CmR, chloramphenicol resistance gene; URA3, orotidine 5-phosphate decarboxylase gene. (C) Table indicating cloning configurations and efficiencies. (D) Linker-mediated recombinational cloning. DNA fragments (100 to 200 ng) plus 20 pmol of oligonucleotides were transformed into MaV203. DNA fragments are represented by red lines, vector by green lines, and oligonucleotides by blue lines.
In Vitro Recombineering
In a parallel approach we sought to accomplish DNA fragment assembly in vitro where the assembled molecules are directly selected in E. coli. For this purpose we developed a highly efficient enzymatic mix that promotes homologous recombination of up to four DNA fragments plus a vector with 15 bp end-homology. (Note: we were able to prove assembly of up to seven fragments, but the mix is optimized for up to four fragments plus vector).
DNA molecules (20 to 500 ng per fragment in a 2:1 insert:vector molar ratio) were incubated with the enzyme for 30 minutes at room temperature, transformed into TOP10 chemically competent cells, and selected on LB agar plates supplemented with the corresponding antibiotic. Cloning efficiencies ranged from 40% to >90% depending on the number and quality of the DNA fragments (Figure 2A).
Recombination can occur not only at the end of the fragments, but it also works at least up to 32 bp away from their ends (Figure 2B). This attribute is useful for generating cloning variants using a single linearized vector.
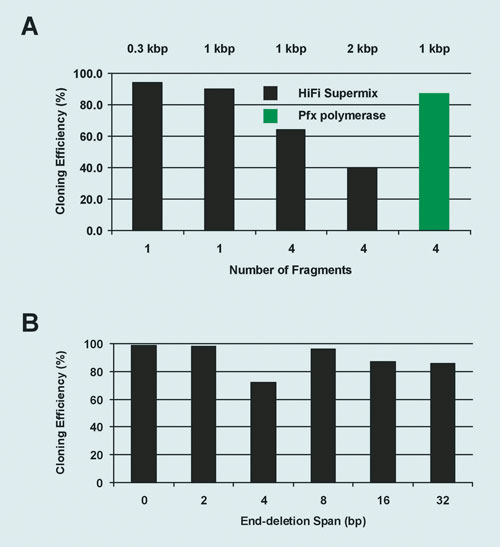
Figure 2. In vitro recombineering: (A) Fragments of the indicated size were amplified using PCR SuperMix HiFi or the proofreading Pfx polymerase and recombined into linearized pUC19. End identity was generated by the addition of a 7 to 15 nucleotide tail to the oligonucleotides. (B) Junction editing: Two DNA fragments were recombined at the indicated distances from their corresponding ends.
The enzyme described above can also be used for recombining and editing the ends of a single DNA molecule, thereby enabling a highly efficient site-directed mutagenesis approach (Figure 3A). Two complementary oligonucleotides with centrally located mutation sites were used.
The DNA methylation and amplification steps were combined into a single reaction. No in vitro digestion or DNA purification was required after methylation or mutagenesis. The system can generate base substitutions, deletions, or insertions of up to 12 nucleotides in plasmids as large as 14 kbp (Figure 3B).
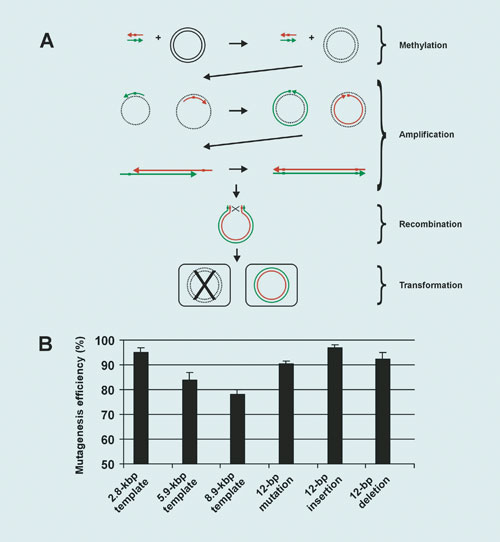
Figure 3. A novel site-directed mutagenesis strategy: (A) Schematic view of the approach. Template strands are shown in black. Methylated strands are shown as dotted lines. Oligonucleotides and new strands are shown in green and red. (B) Plasmids with frameshift mutations in the lacZa gene were subjected to 12 to 18 PCR amplification cycles using a pair of corrective primers, preceded by a 12-minute step at 37ºC. An aliquot was then subjected to recombination for 10 min at room temp, transformed into DH5a competent cells and plated onto LB agar ampicillin X-gal plates. The mutagenesis efficiency is represented by the ratio blue/total colonies. A 14 kbp plasmid was also subjected to mutagenesis. Ten independent random clones were sequenced, and all of them were correctly mutagenized.
Conclusion
In anticipation of an imminent paradigm shift, largely due to the emergence of the synthetic biology field, our ultimate goal is to reach a comprehensive solution to generate any DNA molecule up to high-level genetic systems starting from digital sequences stored in a computer. The approaches presented in this article are relevant not only for the area of synthetic biology but they also have remarkable implications for the current cloning standards.
Lansha Peng is scientist III, Billyana Tsvetanova, Ph.D., and Xiquan Liang, Ph.D., are staff scientists, and Federico Katzen, Ph.D. ([email protected]), is a senior staff scientist, all at at Life Technologies.
Ke Li, Jian-Ping Yang, Ph.D., Tony Ho, Josh Shirley, Liewei Xu, Ph.D., Jason Potter, Wieslaw Kudlicki, Ph.D., and Todd C. Peterson, Ph.D., all of Life Technologies, contributed to the research outlined in this article.
Reagents referenced in this article (Geneart® Seamless Cloning and Assembly Kit, Geneart High-Order Genetic Assembly System, and Geneart Site-Directed Mutagenesis System) are for research use only (not intended for any animal or human therapeutic or diagnostic use). A free on-line tool that facilitates the cloning strategy and generates the required oligonucleotides can be found at www.invitrogen.com/DesignDNAassembly.