July 1, 2011 (Vol. 31, No. 13)
Novel Approach Targets RGS Proteins Using Engineered Gα Proteins and Transcreener® GDP Assay
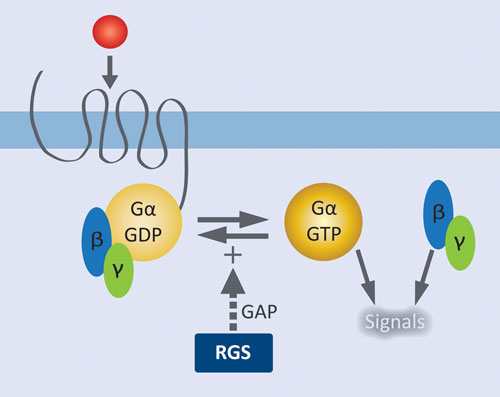
GPCRs are by far the most extensively validated class of therapeutic targets, and there remains tremendous potential for targeting new receptors and their downstream effectors. GPCRs at the cell surface are coupled to a membrane-associated heterotrimeric complex of a GTP-hydrolyzing Gα subunit and a Gβγ dimeric partner. Agonist activation of the GPCR results in release of GDP from Gα and formation of the active complex with GTP. Liberated Gβγ and the activated Gα•GTP complex initiate downstream signaling pathways. Regulator of G protein signaling (RGS) proteins control signal termination by serving as GTPase accelerating proteins (GAPS); i.e., they increase the rate of GTP hydrolysis by Gα.
There are at least 37 RGS proteins encoded by the human genome that contain the signature RGS domain. They are grouped according to sequence homology between their RGS domains and fall into subfamilies with similar multi-domain architectures and selectivity for one or more of the 20 Gα subunits that serve as substrates.
For example, the GAP activity of R7 subfamily members is specific to Gi subunits, whereas that of the GEF subfamily appears specific for G12 subunits. The discovery of RGS proteins and their GAP activity toward Gα subunits resolved apparent timing paradoxes between observed rapid physiological responses mediated in vivo by GPCRs and the slow hydrolysis activity of the cognate G proteins seen in vitro. Moreover, as negative regulators of GPCR signal transduction, the RGS proteins are intriguing potential drug targets, since their inhibition should lead to prolonged signaling from G proteins activated by agonist-bound GPCRs.
Given their role in modulating GPCR signal transduction (Figure 1), it is not surprising that RGS proteins have been implicated as potential drug targets for diverse therapeutic areas, including cancer, cardiovascular disease, inflammation, and CNS disorders. Moreover, there is increasing evidence that they may provide a lever to affect GPCR pathways in defined and specific ways.
Studies using RGS domain-insensitive (RGS-i) mutants of Gα subunits have shown that the actions of endogenous RGS proteins within neurons directly control the timing of GPCR-mediated signaling.
Delayed signal transduction kinetics, as well as increases in agonist potency, are seen upon eliminating the potential for GAP activity by endogenous RGS proteins. The ability to increase GPCR-agonist potency by eliminating RGS-dependent GAP activity shows the untapped potential of RGS inhibitors as potentiators both of existing GPCR-stimulatory drugs and of endogenous agonist-mediated signal transduction.
Evidence is also accumulating that RGS proteins exhibit their effects with selectivity for specific GPCRs. RGS1 is a 1,000-fold more potent inhibitor of the M3 muscarinic receptor than the cholecystokinin receptor, as assessed by agonist-stimulated Ca2+ mobilization in pancreatic acinar cells, despite both receptors having similar coupling to downstream pathways.
Despite their potential as therapeutic targets, there have been very few reports of specific inhibitors and those that have been identified thus far bind covalently. At least part of the reason for this is the lack of robust biochemical assay methods to measure RGS GAP activity. RGS proteins accelerate the rate of Gα-catalyzed GTP hydrolysis by as much as 100-fold, which provides the basis for an in vitro screening assay; moreover both types of proteins are soluble and relatively easy to produce.
However, in the absence of GPCR-mediated nucleotide exchange, it is GDP release (rather than GTP hydrolysis) that is the rate-limiting step in the Gα nucleotide cycle. Thus, to examine the effect of an RGS protein in accelerating GTP hydrolysis by an isolated Gα subunit in vitro, a single round of hydrolysis of radiolabeled GTP is usually performed (a.k.a., the single-turnover GTPase assay).
This standard assay for measuring RGS domain-mediated GAP activity is low-throughput and requires discrete steps of [γ-32P]GTP loading onto Gα, purification of the [γ-32P]GTP-Gα complex, and its immediate use before significant hydrolysis by intrinsic Gα GTPase can occur. The assay also involves isolation (in discrete time intervals) of released [32P]phosphate with activated charcoal precipitation and centrifugation, and finally scintillation counting.
This type of protocol would be very difficult to incorporate into an automated HTS environment. Moreover, measurement of steady-state enzyme activity is the standard approach used for both basic research and HTS; all of the assumptions of Michaelis-Menten kinetic analysis are based on steady-state measurements. Use of a single turnover assay thus adds additional complications in data analysis.
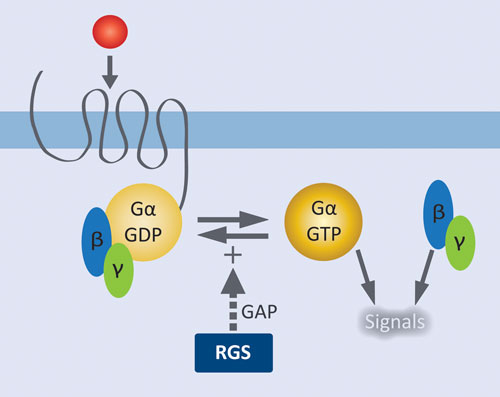
Figure 1. Role of RGS proteins in modulating GPCR signaling: Agonist activation of the GPCR results in release of GDP from Gα and formation of the active complex with GTP. Liberated Gßγ and the activated Gα•GTP complex initiate downstream signaling pathways. RGS proteins facilitate signal termination by increasing the rate of GTP hydrolysis by Gα.
BellBrook Labs’ RGScreen™ is a biochemical HTS assay that detects RGS GAP catalytic activity (Figure 2). There are two key components to the approach: 1) altering the relative rates of Gα GTPase and GDP dissociation so that GDP dissociation is no longer rate limiting allows for the use of steady-state enzymatic assays for monitoring changes in Gα GTPase activity, and 2) selective immunodetection of GDP using the Transcreener® GDP Assay enables homogenous, fluorescence-based detection of Gα GTPase activity in a multiwell format. In combination, these developments enable direct detection of RGS-catalyzed stimulation of Gα GTP hydrolysis in a robust HTS format.
We used well-characterized mutant variants of the Gαi1 and orthologous mutations from related Gα proteins to overcome the disparity between GDP dissociation and GTPase activity. The GTPase activity of the mutated Gα proteins was stimulated six- to tenfold by known interacting RGS protein, demonstrating the ability to directly measure RGS protein GAP activity using steady-state GTP hydrolysis assays.
Importantly, there was no stimulation of GTPase activity by noninteracting RGS domains, demonstrating that the selectivity of the Gα/RGS domain interaction was not altered by the mutations. Moreover, independent binding studies confirmed that the mutated Gα proteins interacted with RGS proteins with the expected specificity.
The double mutant strategy has thus far been successfully applied to three Gα proteins in addition to Gαi1 and also used to measure the GAP activity of four members of the RGS family. We performed a pilot screen of RGS4 protein with a 960 compound library of bioactives in 384-well format. The assay window was more than 100 mP, and the data quality was very high, resulting in a Z’ value of 0.83 and a Z factor or 0.71.
By including control wells containing Gα but lacking RGS protein, we simultaneously counterscreened for Gα inhibitors. Of the 960 compounds in the library, 17 were initially considered hits, and 10 of these were excluded on the basis of the counterscreen resulting in an RGS4-specific hit rate of 0.7%.
More recently, we completed 50,000 compound screens of two RGS proteins with similar assay quality statistics and hit rates.
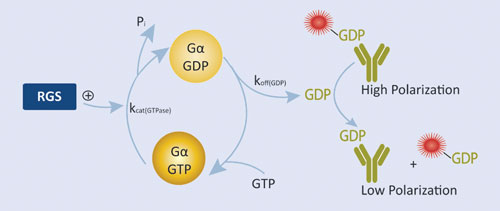
Figure 2. RGScreen Assay relies on proprietary Gα variants with altered GTPase kinetics. Normally, GDP dissociates from isolated Gα proteins very slowly so RGS GAP effects are not detectable using GTPase assays. We have mutated Gαi proteins so that GDP dissociation is no longer rate limiting, yet the proteins still serve as functional substrates in RGS protein GAP reactions. GDP is detected using the Transcreener GDP Assay.
Robert G. Lowery, Ph.D., is CEO of BellBrook Labs. David P. Siderovski, Ph.D. ([email protected]), is professor and director of chemical biology in the department of pharmacology at UNC-Chapel Hill.