June 15, 2012 (Vol. 32, No. 12)
Epitope Mapping and Engineering Complex Membrane Proteins by Comprehensive Mutagenesis
Alanine (Ala) scanning is a widely used mutagenesis approach in which residues in a target protein are systematically substituted for alanine at selected positions by site-directed mutagenesis, expressed, and assayed for function. Substitution with alanine residues eliminates side-chain interactions without altering main-chain conformation or introducing steric or electrostatic effects, so is often the preferred choice for testing the contribution of specific side-chains while preserving native protein structure.
However, because many target proteins are hundreds to thousands of amino acids long, individual alanine replacements across their entire length can be laborious and time-consuming, typically limiting functional analysis to a fraction of the protein’s complete sequence. Furthermore, structurally complex proteins often require eukaryotic machinery for native folding and post-translational modifications, so repeatedly expressing and assaying hundreds of mutations in human cells for each experiment is challenging.
Membrane proteins make up some of the largest and most structurally complex proteins, spanning the lipid bilayer up to dozens of times, forming oligomers, and often existing in multiple conformational states. The human proteome consists of >5,000 membrane proteins, including an estimated 901 GPCRs, 488 transporters, and 237 ion channels, which transmit key information to the interior of the cell and guide important cellular processes such as signaling, transport, and chemosensation.
Their roles in regulating diverse biological processes, their involvement in human diseases, and their exposure on the cell surface has made membrane proteins the targets for >40% of pharmaceutical drugs. Recent high-resolution crystal structures for GPCRs, ion channels, and other structurally complex membrane proteins are beginning to offer new insights into how these proteins function.
Mutagenesis studies such as alanine scanning complement such structures by defining mechanisms of activation and coupling, interactions with ligands and monoclonal antibodies (mAbs), and engineered variants with desired properties.
Shotgun Mutagenesis
Integral Molecular has developed a platform technology, Shotgun Mutagenesis, for comprehensive alanine scanning mutagenesis and high-throughput analysis, enabling epitope mapping and protein engineering even for structurally complex targets that require human cell processing.
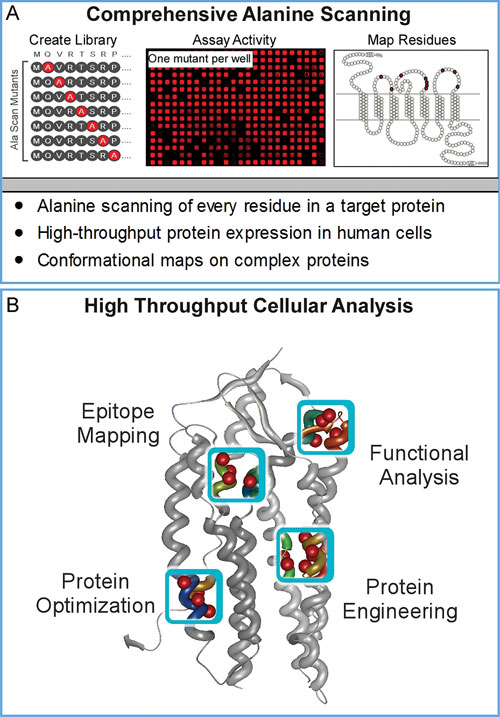
Using automated mutagenesis protocols, every residue in a target protein is changed to alanine (or any other user-defined amino acid) (Figure 1A).
The entire sequence-verified mutation library is arrayed in replicate 384-well microplates for repeated high-throughput expression in live human cells. Each mutation can then be assayed in parallel for its contribution to binding (e.g., mAbs, drugs, or ligands) or for desired protein characteristics (e.g., increased surface expression or solubility) (Figure 1B).
As part of Shotgun Mutagenesis library construction, each mutant clone is validated for conformational integrity (e.g., for function, reactivity with conformational ligands, and/or surface trafficking) to ensure that substitutions do not affect global protein structure. Because the entire process is automated and optimized, Shotgun Mutagenesis enables hundreds to thousands of mutations to be constructed and analyzed in the same amount of time that it would typically require to manually create and analyze several dozen point mutants.
Figure 1. Comprehensive alanine scanning and protein analysis: (A) Each well of a Shotgun Mutagenesis array contains a plasmid clone with an individual, sequence-verified alanine mutation (or other desired residue substitution). 384-well microplate assays are used to assess functional effects of mutants in live human cells and residues of interest are mapped onto protein structures. (B) Shotgun Mutagenesis has been used to map protein interactions at amino acid resolution and to optimize desired protein characteristics.
Epitope and Functional Mapping
Integral Molecular has used Shotgun Mutagenesis to identify epitope maps and functional maps for the GPCR CXCR4, a chemokine receptor that activates cell migration pathways, serves as an HIV co-receptor, and is upregulated in a variety of human cancers. A mutation library of CXCR4 was constructed, individually mutating all 352 residues, and each clone of the CXCR4 library was expressed in live human HEK-293 cells and assayed for binding to a panel of conformation-dependent monoclonal antibodies.
Critical residues that comprise each epitope were mapped onto the recently solved CXCR4 crystal structure to visualize and compare distinct antibody binding sites (Figure 2A). The CXCR4 mutation library was also used to map residues that participate in SDF-dependent signaling by monitoring CXCR4 function (calcium flux) in response to its cognate ligand SDF-1α (Figure 2B). Mutations that mitigate calcium flux help define the structural motifs that make up the SDF-1α binding site and mediate CXCR4 signaling.
Protein Engineering
Integral Molecular is now using its Shotgun Mutagenesis technology to engineer protein variants that demonstrate desired properties such as enhanced expression or solubility. As a proof-of-concept, we screened a comprehensive mutation library of the GPCR TAS2R16, a prototype bitter taste receptor. The TAS2R16 library, comprising mutations at all 291 residues, was expressed in human HEK-293 cells and assayed for both surface expression and total expression by immunoassays.
Comparison of expression measurements enabled the identification of residues in TAS2R16 that affect its surface trafficking, including variants that show significantly enhanced surface expression (Figure 2C, green clones) or decreased surface expression (Figure 2C, red clones).
Protein engineering applications for Shotgun Mutagenesis include identification of stabilized variants. As an example, CXCR4 variants with enhanced stability in solution were identified using an ELISA-based binding assay where human HEK-293 cells expressing the CXCR4 mutation library were solubilized in detergent and tested for antibody binding using a conformation dependent antibody (12G5) (Figure 2D). Stabilized CXCR4 variants allow mapping of the key positions in the receptor that contribute to its detergent stability.
The Shotgun Mutagenesis platform for comprehensive mutagenesis and high-throughput analysis of proteins in human cells facilitates protein mapping and engineering applications previously considered excessively time-consuming or prohibitively expensive, opening discovery pathways for many untapped targets in the human proteome.
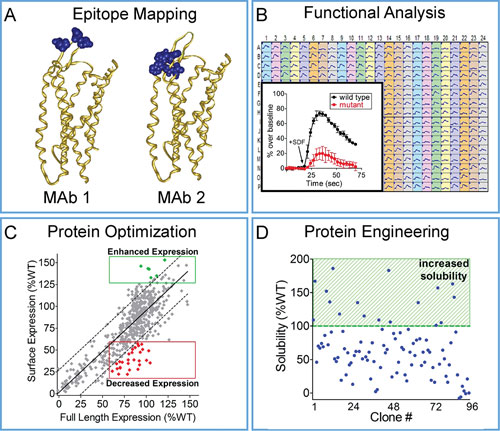
Figure 2. High-throughput mapping and protein engineering: (A) Critical residues comprising distinct epitope maps for two CXCR4 mAbs are visualized on the crystal structure of CXCR4. (B) Critical clones required for CXCR4 signaling show significantly decreased calcium flux (inset, red trace) compared to wild type (inset, black trace) in response to treatment with SDF-1a (HEK-293 cells in 384-well format are expressing each CXCR4 mutant). (C) Critical residues required for TAS2R16 surface trafficking (red clones in lower right) and variants with enhanced surface expression (green clones in upper right) were identified by comparing full-length expression and surface expression for each clone in the TAS2R16 mutation library. (D) To identify variants with increased detergent solubility (highlighted), CXCR4 variants were expressed in HEK-293 cells, solubilized, and probed with the CXCR4-specific conformation-dependent antibody 12G5.
Eli Berdougo, Ph.D. ([email protected]), is a communications scientist, and Benjamin J. Doranz, Ph.D., is president and CSO at Integral Molecular.