April 1, 2016 (Vol. 36, No. 7)
Sara Agee Ph.D. Vice President of Marketing Seegene Technologies
Utilizing the Tagging Oligonucleotide Cleavage and Extension (TOCE) Technique
The real-time polymerase chain reaction( PCR) has become a key tool for molecular assay development, expanding into applications as diverse as in vitro diagnostics (IVDs), food safety testing, and pharmacogenomics. Now, real-time PCR instrumentation systems with multiple fluorescence detection channels have expanded the capacity of PCR to detect multiple analytes in a homogeneous assay, increasing the information gathered per reaction with less sample input.
Multiplex assay development is notorious for presenting certain difficulties, including multiplex probe design, oligo interference, assay design flexibility, and optimization.
To meet challenges such as these, Seegene has developed a new method for high-multiplex real-time PCR technology. This method, which is called Tagging Oligonucleotide Cleavage and Extension (TOCE™), enables detection of multiple targets in a single fluorescence channel through melting temperature analysis of a series of artificial templates. TOCE can enable multiplex assays to be run on existing real-time PCR instrumentation.
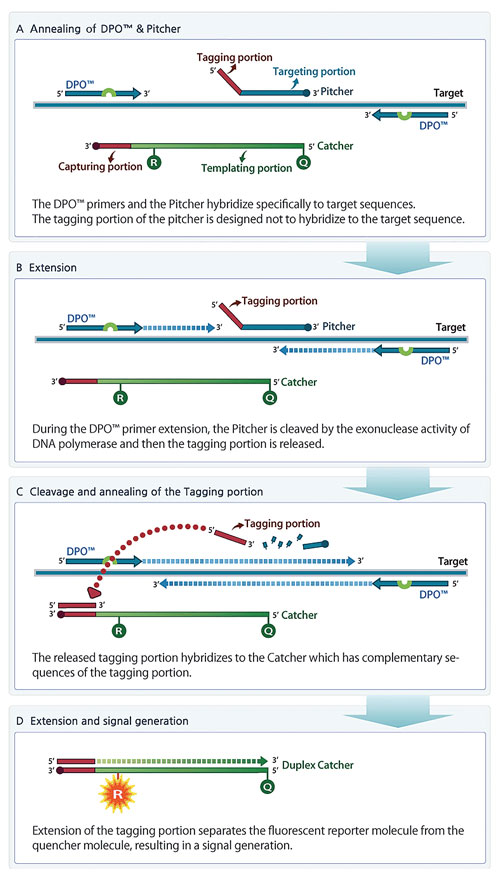
TOCE is a novel approach to real-time PCR and has several unique oligonucleotide components (Figure 1). The key components for TOCE technology are dual priming oligonucleotide (DPO™) primer pairs, “Pitchers” and “Catchers.” The DPO is Seegene’s proprietary target-specific primer design and provides highly specific amplification of the target region. The Pitcher is a tagging oligonucleotide that hybridizes specifically to the target region. The Catcher is a fluorescently labeled artificial template.
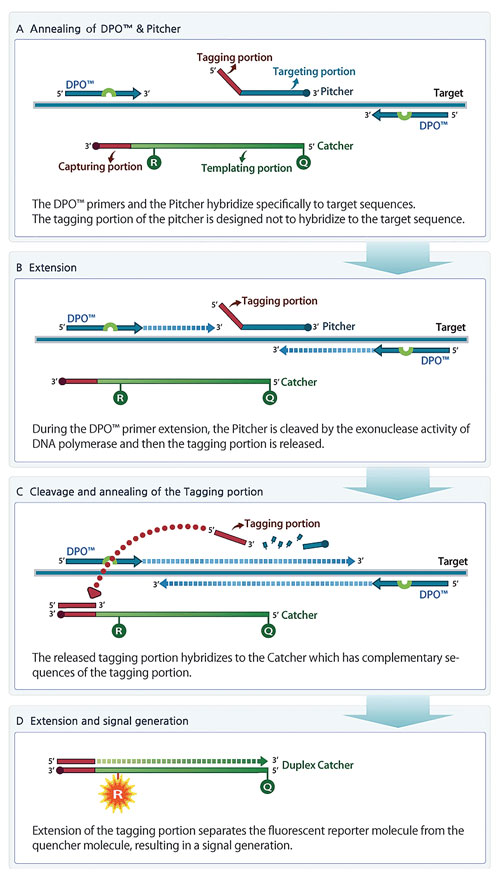
Figure 1. Principles of TOCE assay design and signal generation during real-time PCR. (A) TOCE assay is initiated with hybridization of upstream and downstream primers (DPO) and Pitcher to the selected target sequence. (B) Taq polymerase-mediated extension of primers. (C) Taq polymerase having a 5’ nuclease activity encounters the target-bound Pitcher, and cleaves the Pitcher, releasing the tagging portion. The sequence of released tagging portion is complementary to capturing portion of the Catcher. (D) As the tagging portion is fully extended on the Catcher to create the duplex Catcher, quenching is diminished and the fluorescent signal can be detected.
Enhanced Target Specificity
DPO primers are an essential component for optimizing target specificity of the TOCE assay. They function normally in PCR but increase specificity of target amplification while minimizing nontarget interactions and primer-based artifacts. These attributes are particularly important in a high-multiplex TOCE assay as a means of preventing spurious oligonucleotide interactions that can generate nonspecific signaling from the Catcher, or interfere with the interaction between the Catcher and the tagging portion of the Pitcher.
Ultimately, the use of DPO primers, in combination with the flexibility of TOCE technology, contributes to exceptional ease of use in design and development of TOCE assays. This further distinguishes TOCE from other technologies that require highly sophisticated bioinformatics and extensive target information to provide specificity for but a limited number of targets.
TOCE technology also allows for mutation detection to discriminate even one base difference accurately, enabling detection of clustered point mutations. In Figure 2, a signal is generated only when a mutant-specific pitcher binds to a mutated target sequence, discriminating mutations from a wild-type sequence.
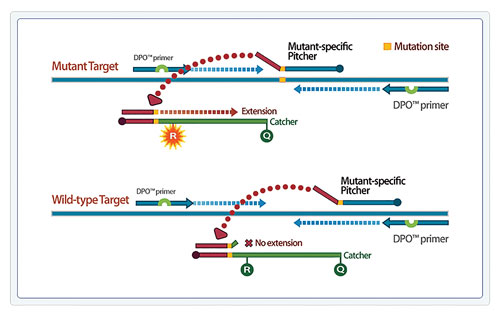
Figure 2. Allelic detection and discrimination enabled by mutant-specific Pitcher-Catcher pairs
High Multiplicity in a Single Channel
TOCE technology enables identification of multiple target analytes simultaneously in a single channel. The signal can be measured in real time and/or analyzed by Catcher melting temperature (Tm) to detect the presence of the target analyte. Figure 3 shows a representative example of Pitcher-Catcher designs to differentiate between three unique melt curves for three targets in a single channel, with Tm values ranging from 60°C, 65°C, and 70°C for the three unique targets, respectively (Figure 3).
Unique target-specific Tm profiles can be controlled by the engineered sequence of TOCE Catcher oligos, and by designing unique paired complementarity of each Catcher to one and only one target-specific Pitcher. One unique feature of TOCE is that the Catcher is a fluorescently labeled artificial template, and therefore the Tm can be controlled by adjusting the sequence design parameters of the Catcher oligo. For TOCE assay optimization, the Tm can be easily adjusted and is not limited by the target sequence.
The controlled Tm of the Catcher oligos result in discrete and predictable melting profiles for each target. As a result, multiple Catchers with unique Tm profiles can be detected in the same reaction and in the same color channel, significantly increasing the number of targets that can be simultaneously detected in the conventional four-channel real-time thermocycler.
One unique benefit of TOCE is the design versatility of the Pitcher-Catcher detection system. Without changing any other component in the assay, the same target could be detected at different Tm values simply by changing the sequence composition and/or length of the templating portion of the Catcher. This controllable Tm property is a key to the ease of design of a TOCE assay. Catcher combinations are flexible, simplifying assay optimization through selection of tagging portion and Catcher combinations to give the desired Tm profile. Moreover, these combinations are interchangeable, so that tagging portions or Catchers can easily be selected to minimize nonspecific interactions.
TOCE is a major advancement in high-multiplex real-time PCR capabilities, offering assay design that greatly expands the capabilities of conventional and next-generation real-time PCR instrumentation platforms. TOCE enables high-multiplex assay design and development across a wide range of applications, including quantitative real-time PCR and highly selective mutational analysis, providing a powerful new tool for high-multiplex molecular assay development in real time.
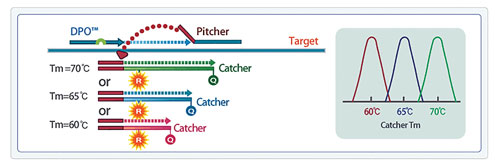
Figure 3. Controlled Tm profiles through designed sequence variations of the Catcher
Sara Agee, Ph.D. ([email protected]), is vice president of marketing at Seegene Technologies.