September 15, 2011 (Vol. 31, No. 16)
Data-Analysis Tool Aims to Skirt Challenges that Have Hampered Technique in Past
Today, label-free is a frequently applied technology. Cellular assays are a major screening application as phenotypic cell changes can be observed without disturbing the native cells. Label-free instruments are used more and more with this application, which creates specific challenges for scientists running experiments. An intrinsic challenge is that the readout of all label-free instruments has no direct link to the biological process occurring in the cell.
While the basic principles of data analysis for such plate-based, time-dependent experiments have been established, most available data-analysis platforms are restrictive when analyzing label-free data. They use only a part of the information content of the traces or require long data-analysis times compared to other screening technologies.
Therefore, label-free analysis can create productivity and time drains and potentially undermine the value of screening lab output. This tutorial addresses how these challenges are overcome with a comprehensive and scalable data-analysis platform such as Genedata Screener® from Genedata.
Specifics of Label-Free Technologies
Label-free sensors measure the changes of cellular phenotypes by specific detection technologies such as impedance, surface plasmon, planar waveguide, or acoustic resonance. Readout changes are related to known and expected changes of the cellular phenotypes such as growth or shrinkage of cells, redistribution of intracellular masses, cellular shape changes, or changing orientation of cells to each other.
A label-free experiment usually starts with measuring the signal of an experimental base status. When stimulating agents (e.g., compounds) are applied to the cells, the resulting phenotypic changes of the cellular phenotypes should cause a signal change. Both the phenotype of the base status (before addition of the compound) and the phenotypic change caused by the compound addition have to be verified external of the instrument (e.g., by verification through a microscope). The signal change corresponds to the difference between these two verified phenotypes.
To better monitor and understand the relation between the signal change and the associated phenotypic change, additional measurements are recorded between the base line and a defined end time. The number and frequency of measurements are determined during assay development based on the confirmed phenotypic changes. Often, there is a specific signal expected between tzero and tend, e.g., a peak indicating a specific temporary, reversible change of the cells. Figure 1 shows example traces from Corning EPIC®.
Simplified (and frequently applied) data-analysis methods just use maximum and base-line measurements. However, the important information in this example lies in the shape of the traces and needs to be used to obtain useful results.
During assay development, experimental conditions and measurement frequencies are defined as well as the data analysis. This includes aggregation of the signal for defined time points (e.g., baseline: mean of first four measurements, peak: maximum signal of time points 12 to 20, etc.). However, during the actual screening experiment, deviations may occur.
- The test substance shows an unexpected effect not captured by the controls used to calibrate the experiment.
- The well shows unexpected issues (e.g., a strong artifact) that dominate the signal of the compound within.
- Signals are shifted from plate to plate—both in strength and time.
These issues are compounded as most screening platforms do not allow analysis of the full-time traces; they only import the aggregated results as defined during assay development. Results are calculated automatically (often in batch mode) by the software accompanying the label-free instrument. This situation has severe data-analysis consequences.
- It becomes next to impossible to see the actual time traces in the context of plate QC or hit-list creation. As a result, important trace trends go undetected reducing a lab’s successful result generation.
- If aggregations must be adapted, data analysis starts from scratch. This requires a parameterization change in the instrument software followed by an export and re-import into the screening software. As a result, one sees inefficient screening, decreased productivity, and the potential for increased error rates.
- When multiple instruments from different vendors are used in the same laboratory, different data aggregation and treatment methods are brought into the lab. As a result, there is inconsistent data processing.
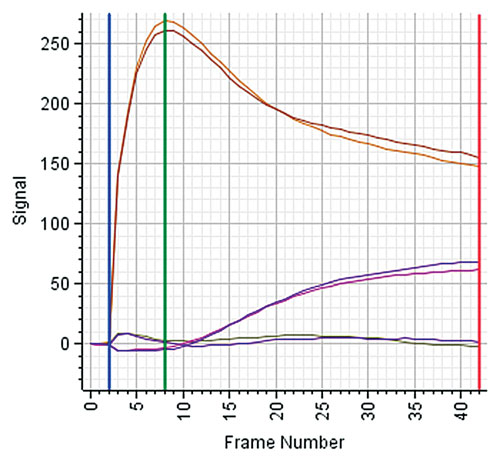
Figure 1. Kinetics traces from a Corning EPIC label-free instrument: There are six traces corresponding to six wells. Two traces correspond to a compound triggering the response of the Gs receptor, two traces to a compound activating the Gi/Gq receptor, and two traces correspond to no treatment (baseline). The trace pairs illustrate the different and characteristic responses of each receptor class: the Gi/Gq receptor shows a fast, yet temporary response reaching a peak at time frame number 8. In contrast, the Gs receptor shows a much slower signal increase, reaching its maximum at time frame 42.
Overcoming These Limitations
Using the Genedata Screener platform, researchers leverage the software’s support for the complete path from kinetics trace to normalized results and hits. With the capability to integrate with widely used SPR-based instruments such as the Octet® (ForteBIO), BIND® (SRU Biosystems) and EPIC (Corning), or impedance-based instruments such as ECIS™ (Applied Biophysics), CellKey™ (MDS Analytical Technologies) and xCELLigence™ (Roche Applied Science), users can adapt data analysis to meet specific screening requirements.
The platform also enables:
- Interactivity: Redefine aggregation rules at any time and immediately see the effect on hit lists, or review kinetics traces of hits. Result: Highly efficient screening, faster hit-to-lead process.
- Scalability: From a single 96-well plate to a full library screen on 300,000 or more compounds, full-time resolution is supported while interactivity is maintained. This allows processing any dataset without dividing it into manageable pieces. Result: Increased productivity and consistent results.
- Instrument independence: Setup and definition of aggregation rules are completely instrument-agnostic, allowing users to set up their own aggregation and QC guidelines, independent of the underlying technology platform. Result: Protection of earlier investments in screening instrumentation and ability to widely share data internally and externally.
Case Study
This example illustrates the analysis of a small compound library screen (10,000 compounds) for two types of GPCR stimulators.
During assay development, the optimal aggregation rules were identified based on the plate quality scores. The data-analysis process consisted of two steps: 1) preprocessing the data from the instrument by the instrument’s software, creating two measurements per well, each thought to be specific to one receptor; and 2) importing this result set twice (one time for each receptor) into a screening data analysis and running the typical steps, normalization, and hit identification for each receptor.
While the results of this experiment convinced the screening lab of label-free benefits, data analysis produced suboptimal results and required lengthy manual inspections of identified hits.
Genedata Screener addresses both of these issues. It offers optimized aggregation rules, utilizing a larger section of the time traces and different methods (e.g., Slope and Area under the Curve). This allows more reliable discrimination between the two receptor types and identification of mixed or new phenotypes.
In addition, Genedata Screener provides integrated data analysis on a single software platform to significantly reduce processing time. The entire library was analyzed for both receptors in a single session in less than ten minutes. Immediate accessibility of the time traces allowed ad hoc comparison of any numerical finding with the underlying original signal.
This enabled the user to approve and combine results with knowledge on the expected phenotypic changes during the data analysis and allowed improved results as the risk to miss an effect is reduced. Figure 2 highlights the effective hit list creation and review.
We increasingly see pharmaceutical companies, specialized contract research organizations, and academic screening centers leveraging the advantages of a single, integrated platform for label-free data analysis. A comprehensive and automated software solution such as Genedata Screener helps to reduce data-analysis cycles, improve screening lab productivity, and help drive optimal data analysis and trace identification.
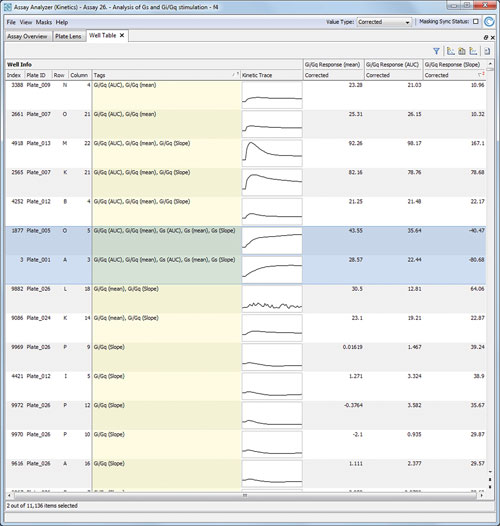
Figure 2. Hit List for Gi/Gq stimulators: Genedata Screener shows the strongest three hits listed here are identified by all three aggregation methods. However, within this result list there is also an ambiguous compound (highlighted, showing ambiguous signature).
Oliver Leven, Ph.D. ([email protected]), is head of professional services at Genedata.



