April 15, 2009 (Vol. 29, No. 8)
Second-Harmonic Generation Provides a Molecular-Level, Functional Readout in Real Time
Conformational change of a biomolecule—a change in its structure—is the ultimate basis for signaling in biology. A convenient and direct way of detecting structural changes in protein targets, and one that is scalable and requires small amounts of sample, would be a big help in definitively identifying compounds that act as agonists or antagonists, and especially useful for discovering allosteric ones. Conventional techniques such as NMR and x-ray crystallography provide atomic-level resolution but are not particularly suitable for drug discovery because of their intensive labor, sample, and time requirements.
This article reviews second-harmonic generation (SHG), a new screening platform for detecting conformational change, that is fast, scalable, and requires tiny amounts of sample. It can be used in either a label or label-free format. It is also suitable for fragment screening up to millimolar concentrations. It provides a direct answer to the question of whether a hit is an antagonist or an agonist, which is often difficult and time-intensive to ascertain with cell-based assays, particularly with fragment screens. In this methodology, protein samples must be purified, and in the case of membrane proteins reconstituted in one of several ways. Because only a picomole of target is needed per well, however, a little protein goes a long way.
As a detection modality, SHG works in a different way than fluorescence. It is a surface-sensitive technique because it only detects molecules right at an interface. Conformational change is detected when biomolecules are immobilized to the surface. Rather than absorbing and re-emitting light as in fluorescence, light is reflected in a nonlinear way off of a surface. The interface (surface plus biomolecules) converts a small amount of red pulsed laser light into second harmonic light at half the wavelength of the incident light.
For example, 800 nm light is converted to 400 nm, a wide spectral difference. Therefore, the shift is in the opposite direction than what is observed in fluorescence. In practice, this makes separating the signal from the incident light easy to do.
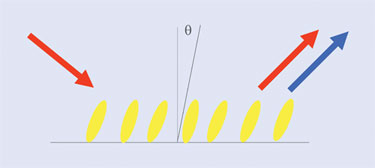
Figure 1. SHG in the physical sciences is used to study molecules on surfaces.
Label and Label-Free Formats
For many years, SHG has been used in the physical sciences to detect molecules and their orientation on surfaces (Figure 1). Only some molecules are SH-active, just as only some are fluorescent. As protein molecules are not generally SH-active, Biodesy has developed both label and label-free formats for detecting targets and their conformational changes on a surface.
For example, in label format purified, labeled protein is immobilized to a surface. A single type of label is used and it does not need to be applied site specifically. Incident light from a laser is directed to the surface. The surface and labels convert a small fraction of the incident light to second harmonic light, the baseline signal.
When the protein changes its structure upon binding ligand or drugs, this changes the amount of second harmonic light produced. In effect, the technique provides an instantaneous and direct readout of conformational change and is thus a molecular-level, functional screen. It is complementary to cell-based assays and can be used in hit generation or the hit-to-lead phase. Only a picomole of protein is needed per assay well. The signal-to-noise ratios are at least 20:1 for a four-second average with z’-factors of about 0.8. The response is in real time. With replicates, one can also measure IC50.
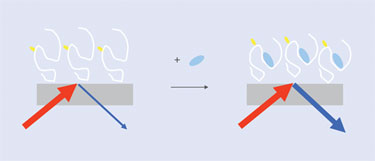
Figure 2. SHG for biomolecule detection in the label format.
Applications
A range of applications are enabled by the platform. One can screen for allosteric compounds directly because SHG detects molecules that block or induce conformational change, not simply those that are competitive at the active site. SHG is also useful for detecting oligomerization and complex formation.
Biodesy is currently offering both services (assay development and screening), as well as a beta instrument for testing and screening. The instrument, Artemis™, is capable of reading 1,000–1,500 compounds per day in wells templated on microscope slides. The slide must be manually changed by a technician after each read cycle. Throughput is more than adequate for focused libraries and fragment screens, and we plan to scale up to HT.
With controls, an SHG assay can provide rapid answers to questions such as: is a compound an agonist or an antagonist; does a compound act allosterically; does a compound stabilize a specific conformation; and does a compound prevent or induce oligomerization or complex formation?
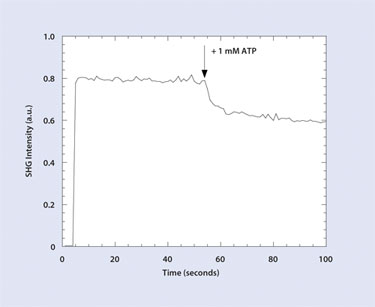
Figure 3. ATP induces a conformational change in adenylate kinase and a change in the baseline SHG signal.
(Produced with permission from the PCCP Owner Societies)
SHG Detection
The general scheme for SHG detection in label format is shown in Figure 2. Data from a typical SHG assay is shown in Figures 3 and 4. Wild-type adenylate kinase, which catalyzes the transfer of a phosphate group from ATP to AMP, is labeled nonsite specifically with an SH-active dye and coupled to a surface.
A baseline signal is generated by the label on the protein. ATP triggers a conformational change and changes the SHG signal. A known inhibitor, AP5A, prevents a subsequent ATP-induced change. The SH signal change can either increase or decrease in a given assay, depending on whether the average orientation of the labels moves toward or away from the surface.
We have used SHG to detect ligand- and drug-induced conformational changes in a variety of protein targets such as amyloids, integrins, enzymes, and membrane receptors. We have also successfully completed several small screening projects for customers.
SHG is a break from incremental advances in drug discovery since it provides a molecular-level, functional readout in real-time. This platform is expected to be a central engine for identifying and validating both conventional and allosteric compounds, at lower cost and in cases where few other methods exist.
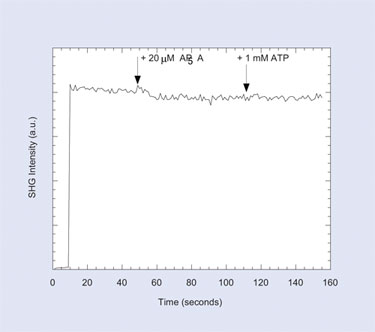
Figure 4. Inhibitor APSA triggers a conformational change when it binds to adenylate kinase and subsequently prevents ATP-induced conformational change.
(Produced with permission from the PCCP Owner Societies)
Joshua Salafsky, Ph.D. ([email protected]), is founder and CSO at Biodesy. Web: www.biodesy.com.



