February 15, 2017 (Vol. 37, No. 4)
Faster, Easier, and More Consistent than Existing Paramagnetic-Based Methods
Recent developments in protein-purification techniques provide the missing tools needed to study weak and transient protein interactions invisible to traditional methods. Complexes that dissociate easily or have short half-lives are lost while they sit in wash buffers and during relatively harsh pipette-based wash steps. But these can be captured with faster and gentler methods, collectively known as Exclusion-based Sample Preparation (ESP™).
ESP is an affinity purification technique that isolates proteins using paramagnetic beads. This unique approach to paramagnetic bead-based isolation pulls analyte-bound beads out of the sample matrix and gently slides them through wash and elution buffers. The surface tension of the liquids gently peels away unwanted debris and the sample spends minimal time in wash buffers, which often promote dissociation of protein complexes. This subtle change significantly reduces carryover, enables isolation of more weak protein interactors, and maintains the remaining samples intact for further study.
The technique could have a range of advantages from basic science to therapeutics. ESP could transform our basic understanding of weaker interactions and their importance in cellular processes, including the interplay of kinases and phosphatases in cell-signaling cascades or the interaction of transcription factors and RNA polymerase in transcription initiation. In the realm of drug development, identifying weak interactions might lead to drugs that can be effective at lower concentrations, possibly alleviating some toxicity issues associated with certain therapies.
How ESP Works
The ESP workflow is relatively simple, as demonstrated by the first commercialized adaption of ESP, Gilson’s EXTRACTMAN®. A sample containing a target analyte is mixed with appropriately coated magnetic beads. A capture magnet attracts the beads, which are then moved through a series of wells. A second, release magnet temporarily disengages the capture magnet, and the beads glide through each wash solution. Ultimately, a final elution (output) well collects the analyte. EXTRACTMAN has four parallel rows of wells that can be used simultaneously for consistent purification. The entire process is completed in seconds, by simply sliding the handle. This process is depicted in Figure 1.
After the paramagnetic particle (PMP)-containing sample is added to a specially designed well-plate, the capture magnet within the upper handle binds the PMPs from the sample. The handle then slides to the next (wash) well, and the lower release magnet slides underneath that well, forcing the release magnet up with the gentle release of the PMPs into the wash well. Next, advancing the release magnet allows the capture magnet to bind the PMPs again and move them to the next step, where the process is repeated through the elution step. The interplay of the two magnets, along with the surface tension of the buffers, allows for the movement of PMPs through the bind, wash, and elute steps of the procedure without disturbing the droplets.
One significant difference between EXTRACTMAN and traditional paramagnetic bead-based procedures is that the paramagnetic bead-bound analyte is moved, rather than the solution. This minor adjustment significantly reduces cross-contamination and the number of steps. ESP does not change the chemistry, but it does eliminate or dramatically reduce the time and steps.
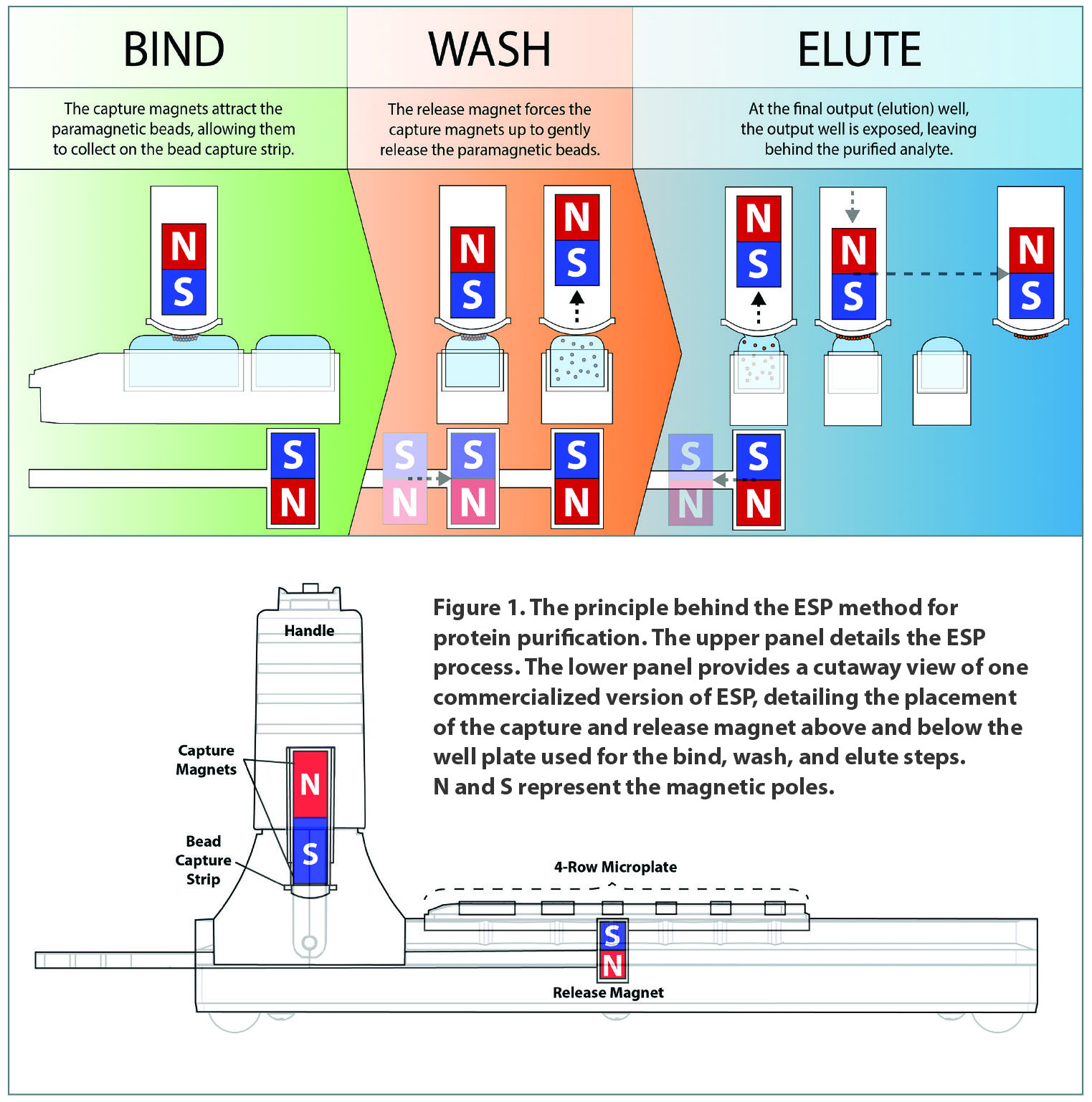
Figure 1. The principle behind the ESP method for protein purification. The upper panel details the ESP process. The lower panel provides a cutaway view of one commercialized version of ESP, detailing the placement of the capture and release magnet above and below the well plate used for the bind, wash, and elute steps. N and S represent the magnetic poles.
Implementing ESP into a Co-immunoprecipitation Protocol
This ESP-based procedure detects more protein-protein interactions than does co-immunoprecipitation (Co-IP), a common technique for studying such interactions. In the example shown in Figure 2, four proteins were used in various Co-IP configurations: Maltose Binding Protein (MBP), Glutathione-S-Transferase (GST), a MBT fusion with Unidentified Protein A (MBP-UPA, and GST fusion with Unidentified Protein B (GST-UPB). The bait and prey protein pairings are given in the Table, and were designed to determine whether the UPA (bait) protein interacted with the UPB (prey) protein. The primary pulldown for the Co-IP reactions used an antibody specific to MBP, bound to paramagnetic beads.
After the Co-IP was complete, the samples were either processed by conventional paramagnetic bead-separation techniques or the ESP-based procedure. Supernatants from both these samples were separated on identical SDS-PAGE gels. Two unprocessed positive controls were included on the gels: GST and GST-UPB. The separated proteins were then transferred to membranes for Western blot analysis using an anti-GST antibody as a probe for the prey protein. The circled band in lane 6 shows the capture of a protein using the ESP method that was not identified using conventional paramagnetic bead-separation techniques.
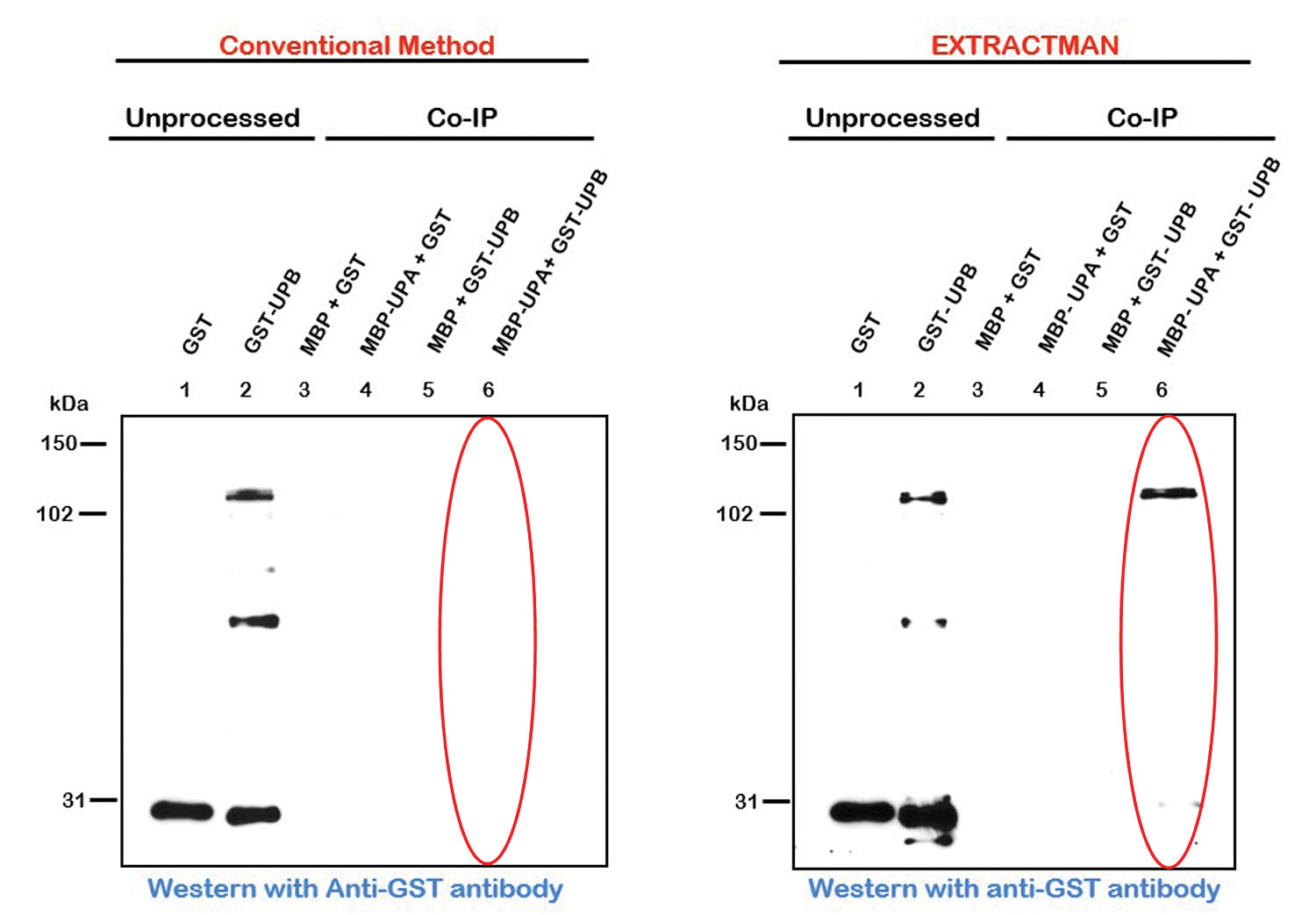
Figure 2. Western blot analyses of GST-tagged proteins following Co-IP by conventional method (left) and with ESP (right).
Why ESP Works
The use of paramagnetic beads made protein isolation faster and easier than earlier methods; however, several issues persisted. First, protein purification remained a bottleneck in protein characterization and identification. Paramagnetic beads were easy to manipulate, but still required a separate tube for each target analyzed. Many times, each experiment called for multiple paramagnetic bead isolations, requiring a larger volume of sample and more time.
A significant issue with the use of paramagnetic beads concerns the multiple wash steps. Traditional paramagnetic bead-based purification methods use numerous, relatively harsh, pipette-based wash manipulations that may result in the loss of weakly-bound or transient protein interactors. Even more problematic is the amount of time the sample spends in wash buffers, which generally promote dissociation. A dissociation constant (Kd) of 10–9 M or tighter typically holds for an hour or so. However, for something with a thousand-times weaker Kd, 10–6 M, the dissociation rate might be about three seconds. Operating the EXTRACTMAN takes as little as 30 seconds, minimizing the time proteins spend in buffers.
ESP offers several advantages. The procedure is fast and easy to use, and is amenable to automation. It also preserves the original sample, which makes reprobing for multiple analytes a possibility. ESP technologies can also purify sufficient protein to perform either mass spec, silver-stained SDS gels, or Western blotting.
Besides these advantages, ESP can use many commercially available paramagnetic beads, making it easy to adapt existing protocols. The ESP principle can also be adapted to allow for multiple protein isolations to be run in parallel for even more speed and sample preparation consistency, and it can be adapted to multiple wash steps.
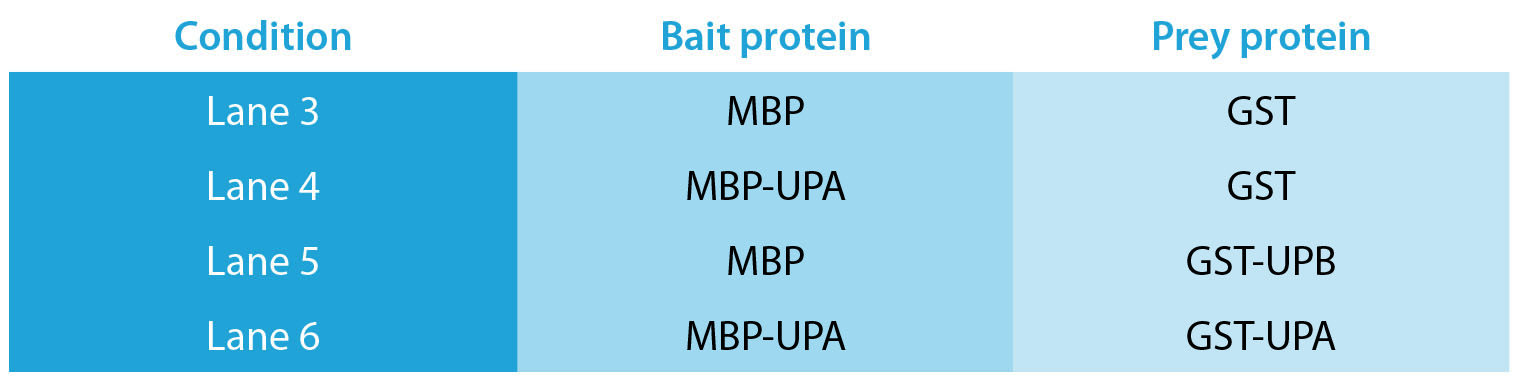
Table. Bait and prey protein pairings for comparison of exclusion-based sample preparation and Co-IP.
Tristan Berto ([email protected]) is product manager of automated liquid handling at Gilson.



