April 1, 2010 (Vol. 30, No. 7)
Improvements in Techniques and Approaches Energize a Once Stagnant Field
Personalized medicine, the application of genomic and molecular data to better target disease states, has been defined as a novel therapeutic concept for over a decade. At CHI’s “Molecular Medicine Tri Conference,” held recently in San Francisco, presenters demonstrated how the early promise of personalized medicine in cancer molecular diagnostics is now being realized.
Tumor model development has three specific challenges—genetic alterations, correct tissue context, and variation—all important in matching up drug response and genetic context. Current xenograft models have variation and alterations but are typically not well characterized. To address these shortcomings, AVEO Pharmaceuticals created a population-based tumor model system based on human-in-mouse tissue transgenic human tumors that feature naturally occurring tumor variation akin to that observed in human tumor populations. The consequence is that tumors vary from one to another, just like human tumors.
“Cell-line derived xenografts have not been good predictors of response in the clinic and, therefore, present big challenges for biomarker discovery,” said Min Wu, Ph.D., principal scientist. “There is a substantial need in oncology for preclinical models that better replicate human cancer. The field is watching closely to see how biomarkers discovered in these new models translate into humans.”
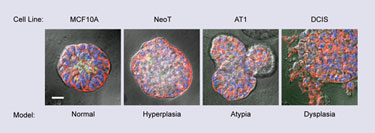
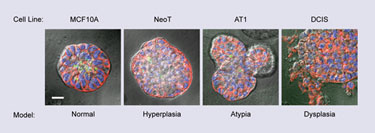
3-D culture models of normal breast structure and premalignant progression to the dysplastic DCIS stage (Wayne State University)
Noninvasive Disease Detection
According to Jack Leonard, president of technology commercialization at febit, the ability to assay all known miRNAs is key to finding valuable cancer biomarkers. “A readily apparent disadvantage for a single biomarker is low sensitivity. One of the best known biomarkers is prostate-specific antigen (PSA). When PSA is present at a typical threshold value of 4 ng/mL of blood, your sensitivity is only 20 percent—which is clearly not a good number because you are only detecting 20 percent of the cancers that are present.”
Historically, Dr. Leonard noted that research has moved forward with many people working on problems independently, typically with one laboratory working with a particular biomarker, while another lab might be working on the same biomarker with a different protocol.
“One problem is that biomarker discovery was not sufficiently standardized, and different laboratories were using different methods,” he explained. “We’ve created an automated approach applying a microfluidic polymerase extension assay (MPEA) for all known human miRNAs and controlling for all the nonbiological variables. We assay biomarkers and samples simultaneously, but hold the other things constant by using the same technology platform. This allows us to do large and well-controlled studies to identify the most informative miRNA signatures.”
Dr. Leonard said that febit’s noninvasive diagnostic assay was specifically designed for the integrated detection of a broad panel of diseases and is well suited for high sample throughput at low cost. “Our sensitivity and specificity for prostate cancer are greater than 95 percent.”
Dr. Leonard also explained the benefits of the Geniom RT Analyzer platform. “We have advantages over other miRNA-expression profiling techniques—the way we manufacture the biochips using light-directed oligonucleotide synthesis, we can turn around new designs quickly. Automation and flexibility are important features of Geniom technology, as this gives clinical researchers the reproducible performance they need to add new results to their growing database on their disease, and the ability to easily change content as more miRNAs are discovered.”
One of the other advantages of the platform is that samples can be collected with minimal discomfort—a blood draw is sufficient to isolate total RNA from the sample and hybridize miRNA onto the biochip. “We hope that this will eventually be used by doctors, but for right now it is more of a clinical research tool—it might be a number of years before this is used as a diagnostic platform.”
Jianfeng Xu, M.D., Dr.PH, professor at Wake Forest University School of Medicine, discussed three types of prostate cancer related to genetic variants that have been found from genome-wide association studies, including those associated with overall prostate cancer risk, aggressive prostate cancer risk, and higher baseline PSA levels.
“We are trying to find genetic markers associated with aggressive cancer,” Dr. Xu said. “The current inability to accurately distinguish risk for life-threatening, aggressive prostate cancer from the overwhelming majority of slow-growing cases creates a treatment dilemma.”
There has been great progress in this field in terms of identifying the SNPs associated with cancer risk, he noted. In the past three years, for instance, 30 SNPs have been identified. And those 30 are consistently validated in studies of European descents.
While researchers, including Dr. Xu’s team, have identified multiple genetic variants associated with the risk of developing prostate cancer in the first place, until now there have been no genetic factors associated with disease aggressiveness. Based on existing evidence that some men are genetically predisposed to developing aggressive prostate cancer, the researchers hypothesized that inherited genetic variants exist that could be used as markers to identify these men at an early, curable stage of disease.
The researchers identified a genetic variant (rs4054823) that was associated with a 25% higher risk of developing aggressive disease. “A single variant with a moderate effect such as this is unlikely to be sufficient on its own at predicting risk for aggressive prostate cancer,” explained Dr. Xu. “But its identification is significant because it indicates that variants predisposing men to aggressive disease exist in the genome. Knowing your risk to prostate cancer is important. But knowing whether you likely develop lethal cancer versus indolent cancer is more important.”

Technician loading processed sample onto a MassARRAY® mass spectrometer (Sequenom Center for Molecular Medicine)
Nucleic Acid Biomarkers
Charles R. Cantor, Ph.D., CSO at Sequenom, explained that oncogenes are the pathways and the targets for next-generation drug development. “The challenge is in somatic mutations,” he said. “They occur in tumor cells, and they do not occur in normal cells at all. Additionally, a tumor biopsy is small and only a small fraction is tumor, so these mutations present at low abundance. Many methods like conventional DNA sequencing cannot find low-abundance mutations and even nextgen is not cost effective to find low-abundance mutations.”
Mass spectrometry, he explained, stands a better chance of picking up these changes because it is sensitive, quantitative, and can detect changes that occur fractionally. Procedures have been developed to enhance the collection of RNA and DNA fragments that enter the peripheral circulation as a result of apoptosis. These include optimized methods of recovering small fragments, amplifying them, and then detecting and quantifying sequence characteristics by nucleic acid mass spec.
Dr. Cantor showcased a customizable commercial product that enables workers in a clinical setting to detect these changes on a larger scale. He noted that the product is used for research purposes by many labs and is sensitive down to about 5%. “We have work ongoing in house that will push sensitivity down to 1 percent or below,” he said. “We currently scan in a research setting 240 mutations in 19 genes and can suggest to a physician what kind of therapies work. It surprises us, but a number of clinicians already want to use it. By the end of the year you will see this in at least three institutions.”
3-D Breast Carcinoma Model
Ray Mattingly, Ph.D., associate professor of pharmacology at Wayne State University, has developed a tractable, in vitro model of ductal carcinoma in situ (DCIS) based on 3-D overlay culture in reconstituted basement membrane.
“Tumors that are the hardest to treat are those that grow slowly but relentlessly,” he said. “Benign breast tumor—ductal carcinoma in situ—is the fourth most common diagnosed cancer in the U.S. It’s hard to study because these lesions are tiny. What we did was to grow a model in a gel, to study and genetically profile.”
Dr. Mattingly’s group has applied and cross-validated whole-genome microarray and digital gene-expression (DGE) analyses to explore the networks and pathways that underlie DCIS. “Not too many people are working on a progression model,” he reported. “We started doing this whole-gene profiling, but we still weren’t getting a full picture.”
DGE analysis revealed a broad range of products that are transcribed outside of standard (NCBI 36.3) genes models. With digital gene expression there’s no preconceived notion of what you are going to find, Dr. Mattingly said. “With DGE, the real work is to get someone who knows what they’re doing to analyze the data. Bioinformatics is an emerging skillset needed in this field, and we are getting an emerging picture of ductal carcinoma that will progress and those lesions that won’t do anything. We are hoping to make this a prognostic tool.”
Targeted Mass Spec
Immunoassays are widely used to measure protein biomarkers in patient blood, but Steven A. Carr, Ph.D., director of proteomics at The Broad Institute of MIT and Harvard, observed that useful antibody reagents do not exist for the vast majority of proteins.
In biomarker discovery today, “thousands of proteins are being identified with often several hundred or more of these observed to be highly differentially abundant between cases and controls. The problem is that there has been no technology, other than immunoassays, to establish if these protein biomarker candidates are detectable in patient blood and that they hold up as potential markers when a few hundred patients are studied,” Dr. Carr noted.
One of the main reasons so few protein diagnostics have been developed is the lack of reagents and technology to build the credentialing information needed, he explained. “Making a new sandwich immunoassay is an expensive and time-consuming proposition, in part, because you need two antibodies per protein candidate to guarantee specificity. Companies are generally unwilling to make an investment in developing reagents for unproven candidate biomarkers.”
In response to this need, Dr. Carr’s team is developing targeted assay methods employing mass spec to screen and quantify low-abundance proteins in plasma. “My lab is using multiple reaction monitoring (MRM) MS as the technology to bridge the gap between discovery and small numbers of proteins worthy of true clinical validation,” he said.
“Peptides derived from each protein candidate of interest are used as surrogates for measuring the change in abundance of the target protein across multiple samples. Quantification is done by spiking synthetic peptides synthesized in a heavy, but stable isotope-labeled form. These peptides are distinguished from the native peptides on the basis of their masses, but they are otherwise identical to the peptides we want to measure.
“The ratio between the observed abundance of the natural peptide and its heavy-labeled counterpart gives us the precise relative quantitative information we require for candidate verification. The peptides we choose to monitor derive from the lists of those we have observed in MS-based proteomic discovery experiments. However, our candidate biomarker proteins also come from the literature and non-MS experimental paradigms such as microarray experiments.
“In these cases, proteomic experimental data may not exist so we use computational approaches to predict which peptides for each protein are likely to give the highest MS response and provide the most sensitive MRM-MS assay. Unlike immunoassays, MRM-MS technology is also highly multiplexible. The method is also far more sensitive than conventional MS methods.
“While we are working hard to develop and apply these methods for biomarker verification in our own studies, another aim is to insure that any lab that would like to use these methods can do so, and that the results obtained will be sufficiently reliable and reproducible. So, a considerable amount of effort is being put into simplifying the process to ultimately allow full automation of sample processing, data acquisition, and analysis, much like clinical measurements are carried out today. ”