August 1, 2017 (Vol. 37, No. 14)
MaryAnn Labant
Improved Cancer Immunodiagnostics and Immunotherapeutics Depend on Biomarker Discovery
Biomarkers are routinely measured and evaluated as indicators of normal biological processes, pathogenic processes, or pharmacological responses to a therapeutic intervention. Concomitant with the discovery and use of increasing numbers of biomarkers is the technological need to detect these multitudes of proteins accurately and with increased sensitivity. Numerous emerging techniques are focused on facilitating this process.
In May, Cambridge Healthtech Institute’s 13th Annual Biomarkers and Immuno-Oncology World Congress brought together industry experts to discuss advancements to drive innovation and collaboration in the rapidly developing field of biomarkers, diagnostics, and immunotherapy.
Multiplexing in immuno-oncology is increasing. This trend, however, imposes new requirements. For example, novel markers and marker signatures will be useful in the stratification of drug responders vs. nonresponders only if they provide sufficient accuracy. Also, combination therapies and less-invasive biopsy procedures will require more efficient analytical tools to extract more content-rich information from fewer and smaller samples.
Preserving Spatial Information
Multiple cell subpopulations define the tumor microenvironment, such as tumor cells, tumor-infiltrating lymphocytes, and stromal cells. Specific biomarkers are used to detect these cell populations and their spatial relationships to understand the biological mechanisms underlying response to immunotherapy; numerous biomarkers may need to be imaged on single tumor-biopsy sections.
Commonly used, spectral-multiplexing methods are based on successive iterations of antigen retrieval, staining, and amplification steps. Multiplexing often results in long cycle times per sample and likely causes irreversible damage to the tissue sample’s integrity and antigenicity.
According to Louis Levy, director, corporate and business development, Ultivue InSituPlex is a flexible immunofluorescence platform used for the multiplex detection and analysis of protein markers in tissue using stable DNA–DNA interactions. Measurements are order independent and allow high multiplexing levels with fast cycles without tissue or antigenic damage.
Complementary strands of DNA are associated with either an antibody (to bind to a target) or a fluorophore (to image). Multiple targets are stained simultaneously in a single staining step using antibody-DNA conjugates, where each conjugate contains a target-specific DNA strand serving as a barcode.
The barcodes are extended through a single DNA-based amplification step to bind to complementary fluorophore-labeled DNA strands and image a target of low- to high-level abundance. The DNA strands can be dehybridized to completely remove the fluorescence signal from the sample, if desired.
InSituPlex Discovery relies on sequential multiplexing to image double-digit numbers of protein markers. Multiple cycles of hybridization, imaging, and dehybridization are carried out on single formalin-fixed paraffin-embedded (FFPE) sections. After completion of image acquisition, the resulting images are digitally overlaid to visualize the targets of interest using available software packages (i.e., HALO, Visiopharm, and ImageJ).

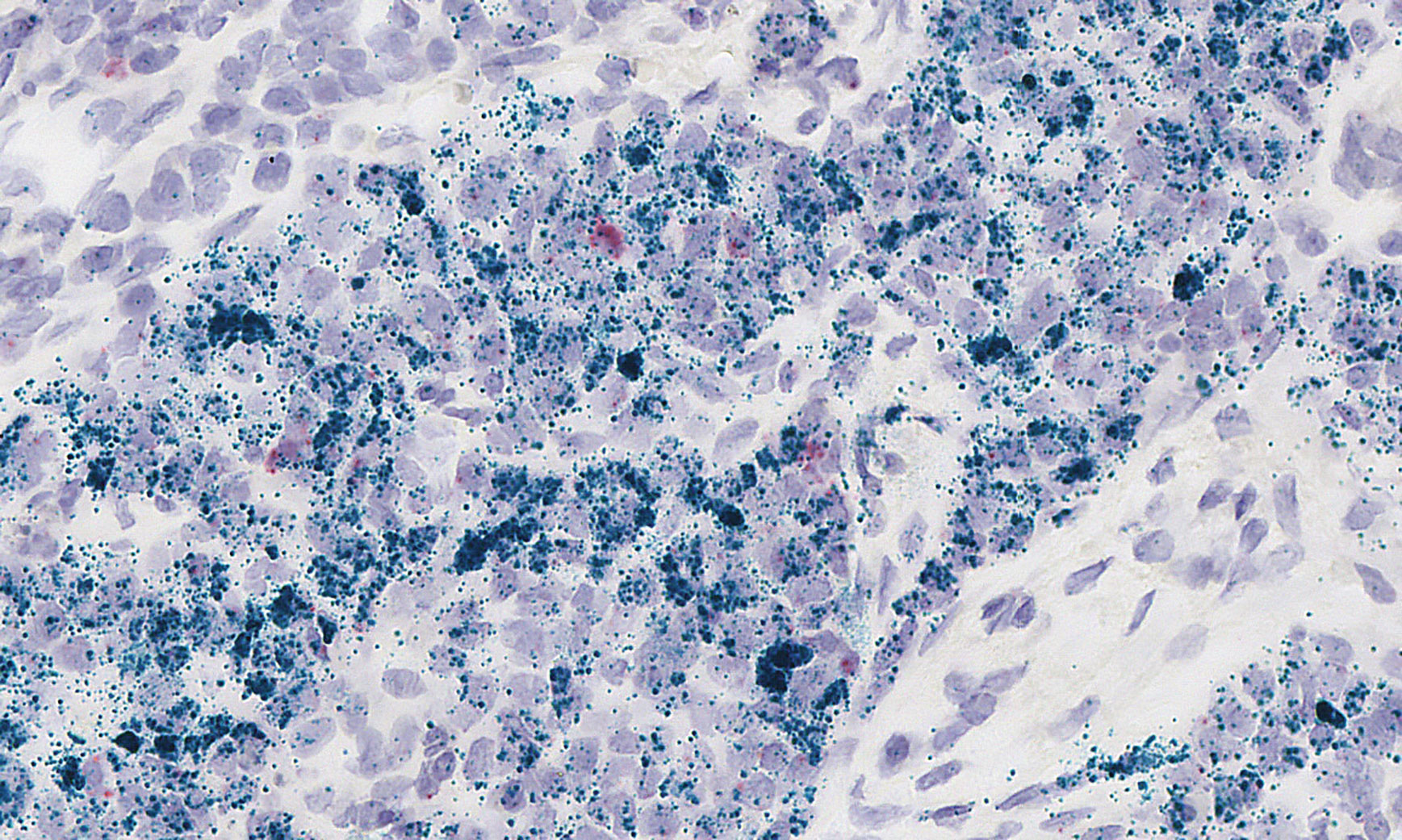
Ultivue has developed InSituPlex, a platform that uses stable DNA-DNA interactions to sequentially image targets. Multiple cycles of hybridization, imaging, and dehybridization are carried out to achieve high multiplexing in situ. In these images, which show human tonsil tissue, the Ultivue-2 two-plex kit for super-resolution microscopy has been used to accomplish six consecutive cycles of addition and removal of fluorophore-labeled DNA strands. The gentle removal of fluorescence in the Ultivue-2 workflow helps maintain tissue integrity.
Quantitative RNA ISH
Immunohistochemistry (IHC) and RNA hybridization (ISH) are widely used to analyze biomarkers at the tissue or single-cell level while preserving the morphological context. These techniques have been incorporated into RNAscope, an ISH assay developed by Advanced Cell Diagnostics for the detection of target RNA within FFPE or fresh frozen tissues.
The assay has a proprietary probe design to amplify target-specific signals, but not background noise, from nonspecific hybridization; two independent probes (double-Z) need to hybridize to the target sequence in tandem for signal amplification to occur.
Because it is highly unlikely that two independent probes will hybridize to a nonspecific target right next to each other, this design ensures selective amplification of target-specific signals. For each target RNA species, 3–20 double-Z probe pairs are designed to specifically hybridize to the target molecule region.
“The absence of universally accepted standards and guidelines for antibody characterization, including target sensitivity and specificity, limits the confidence of IHC data and supports the need for alternative biomarker validation methods,” says Rob Monroe, M.D., Ph.D., CMO, Advanced Cell Diagnostics.
“With fast development timelines, RNA ISH is an excellent method for validation of biomarkers discovered through a variety of genomic approaches, such as DNA-seq, RNA-seq, digital PCR, and microarray, as well as for the validation of antibodies used for detection of biomarkers with IHC,” adds Dr. Monroe. “The RNAscope technology’s unique probe design strategy, which simultaneously amplifies signal while suppressing background, allows for routine and reliable in situ detection at single-molecule sensitivity.”
RNAscope is available in fluorescence as well as chromogenic singleplex and multiplex assay formats. Automated on both the Leica Biosystems Bond platform and the Ventana Medical Systems Discovery platform, the half-day turnkey assay provides consistent, easy-to-interpret, and quantifiable results.
Approximately 15,000 off-the-shelf probe targets are available; new targets can be developed within two weeks. Pharma assay services are also offered for target validation, target safety assessment, and biomarker discovery to support preclinical research, clinical biomarker, and companion diagnostic assay development.
Inflammatory biomarkers, such as secreted factors and their receptors, may be detected with RNAscope in situ hybridization assay technology provided by Advanced Cell Diagnostics. The technology, which has diverse applications (including neuroscience, stem cell, and gene-therapy applications), enabled researchers to produce this image, which highlights the RNA expression of IDO1 (green) and IFNG (red) genes in human lung cancer tissue.
Ultra-Sensitive Protein Measurement
Simoa, another new technology, uses single-molecule measurements to assess previously undetectable proteins, or those only observed in acute responses, in blood across a variety of therapeutic areas, such as neurology, oncology, cardiology, infectious disease, and inflammation.
The technology isolates individual immunocomplexes on paramagnetic beads using standard ELISA reagents. Molecules are trapped in femtoliter-sized wells, allowing a “digital” readout of each individual bead to determine whether or not it is bound to the target analyte.
“The digital feature allows a 1,000× sensitivity increase with coefficients of variation of less than 10%. The device is so sensitive that it can find a grain of sand in 2,000 Olympic swimming pools and find a single blade of grass in a field as big as the state of Alaska,” explains Kevin Hrusovsky, executive chairman and CEO, Quanterix.
For example, Simoa assays can detect neurodegeneration markers in serum and plasma that were previously only detectable in cerebrospinal fluid samples, and can directly detect critical biomarkers for measurement of latent replication competent viral reservoirs, such as interferon-α in serum and HIV p24 protein from single cells.
“Precision biomarker measurement is crucial to realizing the potential of precision medicine. Monitoring biomarker baseline levels is becoming a key element of early detection and treatment approaches. It is our hope that one day clinicians will use technology like ours and a simple blood test as part of annual physicals, to create a baseline patient-specific protein profile,” continues Mr. Hrusovsky.
Recently, the company introduced the Simoa Neurology 4-Plex A assay (N4PA), the first comprehensive multiplex panel to study traumatic brain injury and other neurogenerative conditions. N4PA simultaneously measures four protein biomarkers from either cerebrospinal fluid or directly from blood samples.
A planned benchtop Simoa instrument will support increased levels of multiplexing and provide the ability to measure circulating protein and nucleic acid biomarkers on the same platform. In addition, the Quanterix Accelerator Lab provides dedicated laboratory resources to facilitate new biomarker research, customer assay development, and clinical sample testing.
Monitoring Cell Proliferation
Many biomarkers, such as protein or genetic signatures, indicate the likelihood that a malignancy will respond to a therapy, but do not provide information as to how fast the malignancy is growing, also known as the proliferation rate. Similarly, many traditional serum biomarkers, such as CEA, PSA, and CA15-3, provide information regarding tumor mass and past growth but not the current or the future expected growth rate.
The AroCell TK 210 ELISA is a novel, sensitive, and specific assay for serum thymidine kinase 1 (TK1), a key enzyme in DNA replication and a well-known cell-proliferation biomarker. The assay offers the potential to monitor cell proliferation from a standard serum sample. Most other proliferation biomarkers, such as KI-67, are immunohistological tests performed on tissue biopsies.
“Proliferation biomarkers tell you how fast a tumor is growing, so they are forward-looking biomarkers,” discusses Martin Shaw, business development manager, AroCell. “The TK1/TK 210 protein can be used to study a wide range of malignancies and is a potentially valuable complementary diagnostic for studying therapeutic effects and as an aid in making prognoses. Combining TK1/TK 210 with other biomarkers may lead to increased diagnostic accuracy plus better information to evaluate therapeutic responses in clinical trials.”
Serum TK1 enzyme activity measurements have been used as a cell-proliferation biomarker for close to four decades, particularly in hematological malignancies. However, the measurement methods are complex, and require specialized equipment.
Staffan Eriksson, M.D., Ph.D., professor, Swedish University of Agricultural Sciences, and an AroCell cofounder, developed TK1 specific monoclonal antibodies based on the TK 210 epitope. Studies using these antibodies showed that TK1 is found in serum in high molecular-weight complexes and much is enzymatically inactive, particularly in subjects with solid tumors.
The AroCell TK 210 ELISA assay procedure includes a proprietary sample dilution buffer that releases TK1 protein from these complexes, enabling the quantitative measurement of serum TK1 protein levels. A standard microtiter plate ELISA, the AroCell TK 210 ELISA, demonstrates similar performance characteristics to the reference TK1 enzyme activity assays in hematological malignancies but is, in most cases, much more sensitive in measuring serum TK1 from subjects with solid tumors. The CE-marked assay is currently for research use only.
Visual Analytics
The exploration and analysis of aggregated datasets is often not possible, because the biomarker discovery analytic tools are, in most cases, unavailable. Consequently, insufficient robust novel biomarkers are identified for further validation.
Once different data types associated to a translational project are aggregated, data-analysis challenges can arise. These range from the identification, as well as elimination, of batch effects that can mask the real biological effects; the discovery of the correlations between measurements and observations, such as cytokines changes and adverse events; using robust statistical methods; and the validation of the potential biomarkers.
“PerkinElmer Signals for Translational addresses biomarker-discovery challenges by combining a cloud-based platform for collaborative translational project data management, with a best-in-class visual analytics environment powered by pertinent statistical methods and embedded as agile TIBCO Spotfire Apps,” comments Eduardo Gonzalez, Ph.D., product manager, informatics, PerkinElmer.
The Translational apps allow for the detection and removal of batch effects, identifying, ranking, and visualizing correlations in aggregated datasets with clear metrics to maximize the probability of biomarker validation. Statisticians and bioinformaticians can extend the collection of apps to align with a project’s specific needs.
The objective is to empower scientists with embedded, well-established analytics, allowing the direct exploration and analysis of complex datasets—including omics data—to progress biomarker discovery projects.
There is a strong need for domain experts, like oncologists, to directly gain insight by exploring and analyzing complex translational datasets in a rich visual environment. Advanced analytics for biomarker discovery should facilitate this self-service data exploration and analysis as well as manage streaming data from sensors in hospitals, at home, or even on wearable devices, which is arguably the next domain challenge.



