September 1, 2013 (Vol. 33, No. 15)
Detecting Tumor Mutations in Peripheral Blood Using Digital PCR
Highly specific biomarkers that provide information about cancer initiation, development, metastasis, and response to therapy have the potential to offer prognostic insight, while also facilitating the design and implementation of targeted treatments.
Because of the genetic complexity and diversity of cancer, these biomarkers are most useful when they identify a specific group of patients that would benefit from a highly targeted treatment, specifically optimized for their disease subtype. However, this is further complicated by the continued accrual of somatic mutations over time, for example, those driven by selective pressures exerted by the host immune system and external drug treatments. Therefore, it is necessary to regularly evaluate the biomarker signature of a cancer for changes over the course of treatment and during remission.
Cataloguing the genetic status of a given tumor traditionally involves carrying out sequencing analysis of fresh and/or archival biopsy specimens. However, there are several limitations to this approach. For example, tumor biopsies are often contaminated with the DNA of normal somatic cells, while the cancer cell population within a tumor tends to be genetically heterogeneous in nature.
The use of archival tissue samples, particularly if fixed and embedded in paraffin, can introduce further sources of error due to DNA degradation, which can interfere with analysis. Lastly, tumors can often develop in areas of the body difficult or impossible to access surgically, and can therefore not be assessed using this approach.
Noninvasive Biomarker Analysis Using Peripheral Blood
Fortunately, rapid developments in the technology and protocols required for reliably isolating and analyzing circulating tumor DNA (ctDNA) in peripheral blood are making it easier to obtain the necessary samples without the need for physical biopsies. One such method, developed by the team at Inostics, utilizes digital PCR to detect somatic tumor mutations in ctDNA with high sensitivity. The technique combines emulsion digital PCR and flow cytometry. Known as BEAMing, it employs a combination of beads, emulsification, amplification, and magnetics to achieve the necessary level of sensitivity.
The BEAMing process (Figure 1) starts with isolating and purifying the DNA present in blood plasma, which then goes through a pre-amplification step using conventional PCR to amplify the genetic section of interest using primers incorporating known tag sequences. These DNA templates are then amplified again via emulsion PCR, employing primers directed at these sequence tags and covalently bound to magnetic microbeads via streptavidin–biotin interactions. By designing the system this way, the PCR products generated in each emulsion droplet will remain physically affixed to the microbeads at the end of the reaction, allowing them to be easily separated and purified using a magnet.
The method also provides a digital readout of copy number, as it has been experimentally optimized to ensure that each emulsion droplet will contain a maximum of one microbead and one DNA template molecule. This makes it possible to detect even very rare mutant templates at copy ratios greater than 1:10,000.
Following the emulsion PCR reaction and subsequent purification, the DNA attached to the beads is denatured and probed to determine the presence and number of known mutant variations. Any number of a range of fluorescence-based methods for distinguishing two alleles can be utilized for this step, as long as beads housing the wild-type or mutant DNA can be easily differentiated using flow cytometry.
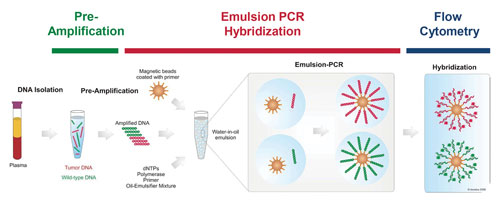
Figure 1. The key steps of BEAMing digital PCR analysis
Importance of Accurate Amplification
The number of ctDNA copies circulating in the peripheral blood stream is very low, especially in comparison to wild-type DNA. For this reason, DNA amplification using PCR is an important part of the BEAMing process, necessary to ensure reliable detection. However, PCR is not an error-free reaction, and any experimental variability introduced by the process increases the risk of false-positive detection, thereby reducing the effectiveness of the diagnostic test. This risk can be minimized by using a high-fidelity DNA polymerase such as Thermo Scientific™ Phusion™ High-Fidelity DNA Polymerase, an engineered enzyme containing a DNA-binding domain fused to a Pyrococcus-like proofreading polymerase.
Because of this unique fusion technology, Phusion DNA Polymerase generates PCR products with a level of accuracy and speed unattainable with a single enzyme, even for GC-rich templates. It also has extremely high fidelity, with an error rate approximately 50-fold lower than that of Taq DNA polymerase and 6-fold lower than that of Pfu DNA polymerase, as determined by a modified lacI-based method (Figure 2) and later confirmed using Roche/454 and Illumina sequencing.
By using a DNA polymerase with such a low error-rate, one can be highly confident that mutations detected by BEAMing are reflective of the original ctDNA template, rather than introduced by an error-prone DNA polymerase.
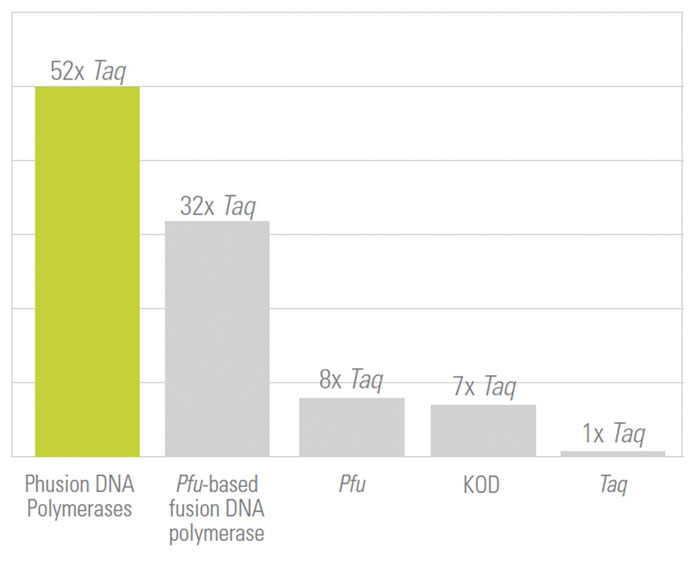
Figure 2. The Phusion DNA polymerase is a hybrid enzyme that delivers high PCR fidelity (relative fidelity of different DNA polymerases shown, where fidelity = 1/error rate).
BEAMing in Action: Detecting PIK3CA Mutations in ctDNA
Recently, a research team led by Ben H. Park, M.D., Ph.D., of the Johns Hopkins Sidney Kimmel Comprehensive Cancer Center and including scientists from Inostics worked to assess the suitability of BEAMing for detecting PIK3CA mutations in ctDNA, using plasma samples from patients with metastatic breast cancer. PIK3CA is the most commonly mutated oncogene in breast cancer, and aberrant PI3K pathway signaling is being actively studied as a prognostic marker for disease outcome and a predictive marker for developing and administering highly targeted therapies.
Part of the study involved using BEAMing to perform a retrospective screen of 49 patients with metastatic breast cancer. The aim was to use the new technique to detect PIK3CA mutations in matched ctDNA and archival tumor samples, to assess whether blood samples might replace biopsies as a reliable method for assessing tumor genotype.
Of the 49 patients tested, 14 exhibited a PIK3CA mutation (Table). In every case, the result obtained using DNA from the biopsy matched that using the corresponding ctDNA sample, indicating that BEAMing of ctDNA is a reliable method for assessing tumor genotype without the need for a biopsy. With multiple inhibitors of phosphoinositide 3-kinases currently in development, BEAMing has clear applications for clinical trial design and execution, as well as future clinical treatments designed to target subsets of patients exhibiting distinct tumor genotypes.
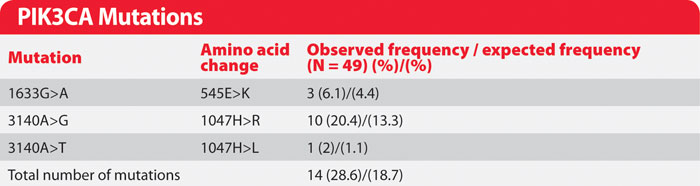
Table. PIK3CA mutations identified retrospectively in peripheral blood of patients with breast cancer. Note: the BEAMing of ctDNA and breast tumor tissue yielded results with 100% concordance (full dataset originally published by Higgins et al., 2012).
Conclusions
BEAMing is a powerful method for detecting known genetic mutations in a blood sample, even when at very low copy numbers. It is much more sensitive than most competing techniques, while also enabling copy-number quantification. The accuracy of the method relies upon the utilization of high-fidelity DNA polymerases, which minimize error rate and ensure that the detection of false positives/negatives does not significantly lower the diagnostic value of the technique. By enabling the analysis of ctDNA via noninvasive biopsies such as systemic blood samples, BEAMing has many applications including optimizing patient recruitment during clinical trials.
In the future, BEAMing may also prove valuable as a diagnostic technique for making informed treatment decisions, as well as for tracking the occurrence of resistance mutations during and after treatment, so that medication can be adjusted accordingly to maximize therapy effectiveness.
Frank Diehl is CSO and managing director at Inostics, and Edita Smergeliene ([email protected]) is product portfolio manager for gene detection at Thermo Fisher Scientific.



