November 1, 2009 (Vol. 29, No. 19)
Premier Biosoft’s AlleleID Was Designed to Assist in Development of Diagnostic Assays
Efficient clinical diagnosis of pathogens is important for the management of infectious diseases. Conventional methods have longer turnaround time and, in most cases, lower sensitivity. Nucleic acid based methods for detection of microorganisms are rapid, sensitive, and are generally successful even when the culturing of microorganisms fails. Sequence-based molecular methods such as real-time PCR provide rapid diagnostics and higher sensitivity, allowing differentiation between related strains.
Although numerous technologies are available for the molecular level identification of pathogens, the most common is real-time PCR. This technique has a high accuracy when used with specific primers and probes. Manual design of these primer and probes is tedious and results in lower quality results owing to the inability to simultaneously handle multiple criteria for design. AlleleID® from Premier Biosoft is a new tool that designs real-time PCR assays to identify and detect pathogens.
Oral pathogens such as periodontopathic bacteria are the etiological factors in the development of gum diseases. Quantitative analysis with the identification of periodontopathic bacteria is important for the diagnosis, therapeutic evaluation, and the risk assessment of periodontal diseases. Real-time PCR is often used for this process. Scientists at Premier Biosoft have quantified six—Porphyromonas gingivalis, Tannerella forsythia, Actinobacillus actinomycetemcomitans, Treponema denticola, Prevotella intermedia, and Prevotella nigrescens—using real-time PCR.
In our recent studies, plaque and tongue debris specimens were collected from 10 patients with advanced periodontitis and 10 periodontally healthy individuals. All species, except for P. nigrescens, were detected in diseased samples in significantly greater numbers than in those from healthy patients, which had greater number of P. nigrescens.
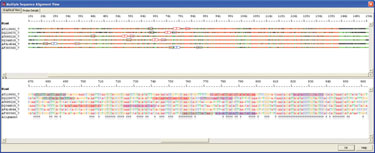
Sensitivity of real-time PCR to detect periodontopathic bacteria depends on the ability of the species-specific primers and probes to amplify and detect all possible species circulating in the population. An example of species-specific detection is shown in Figure 1. 16s ribosomal RNA gene sequences from six periodontal bacteria were obtained from GenBank and aligned using AlleleID. The unique regions identified were used for the assay.
Species-specific TaqMan® probes were designed to detect six bacterial species. These primer-probe sets uniquely detect each of them in the sample. The sets using AlleleID gave an efficiency of more than 93.5%.
In cases where the nature of the sequences is such that a unique probe for each strain cannot be designed, a minimal set using fewer probes can be used to detect all the sequences in an aligned set. Such an assay is useful for for major histocompatibility (MHC) proteins. They are the cell-surface antigens present in most eukaryotes that identify a particular tissue type. In humans, MHC typing plays an important role in tissue grafting.
Due to high conservancy, minor differences in the MHC sequences of different species were observed. In many cases, each sequence was identified with a unique probe without cross hybridizing with any other sequence in the alignment. If such a design fails, the minimal probe set design option can be used. Though a single probe can identify more than one sequence, the combination of such probes uniquely identifies a given sequence.
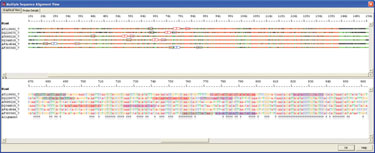
Figure 1. Alignment of six 16s ribosomal RNA gene sequence of periodontal bacteria
Taxa-Specific Detection
Rotavirus is one of nine genera in the Reoviridae family. Based on the antigenic properties of the VP6 protein, the rotavirus genus is divided into different serogroups (labeled A through E with two possible additional species F and G).
Rapid and early identification of the pathogen is highly important for the diagnosis of the disease. Although the existence of varied strains of the virus is established worldwide, certain regions in their genes are conserved. These regions are used for identification of the virus during diagnosis.
Real-time PCR detection assays commonly target VP7 using consensus primers and probes designed to detect all subtypes. In this case study, we evaluated 100 fluid stool specimen collected from children less than five years old suffering from acute diarrhea. All patients were subjected to thorough history taking and clinical examination at the time of specimen collection. Data regarding the age, sex, residence, duration of diarrhea and frequency of motions per day, the presence or absence of fever, vomiting and flu-like symptoms, stool consistency, and breastfeeding were obtained.
Initially VP7 glycoprotein genes of different human rotaviruses genotypes were obtained from GenBank and aligned using AlleleID. The samples included only human rotaviruses of all known G genotypes. The detection of target VP7 glycoprotein gene for 12 strains was performed using TaqMan probes designed in the conserved region to identify presence of any of the known strain of virus in the sample (Figure 2).
In an attempt to further check the specificity of designed oligos, they were BLAST searched against custom databases (experimental sequences used to design species specific assay) created on a local server. No homologies were found indicating their high specificity.
Pathogen-detection and -identification assay design has always been a challenge. Traditional methods can detect and identify pathogens, however they are time consuming and often have poor accuracy. Sequence-based assays for detecting and identifing pathogens are rapid, have high sensitivity, and high accuracy. The design of primers and probes for pathogen identification requires sophisticated software capable of handling multiple sequences, database cross homology identification, and primer/probe thermodynamic considerations simultaneously.
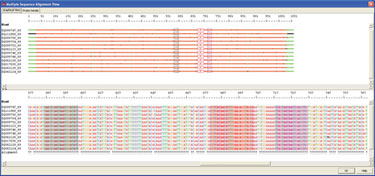
Figure 2. Alignment of 12 rotavirus strains
Arun Apte ([email protected]) is the business development manager at Premier Biosoft International. Web: www.premierbiosoft.com. TaqMan® is a registered trademark of Roche Molecular Systems. SYBR® is a registered trademark of Molecular Probes.