Going the full distance is the least any messenger can do. A messenger should also retain enough vigor to ensure that its message is heard and, if possible, heeded. For an inspirational example, look at Pheidippides, the first marathoner.
Although he ran to the absolute limit of his endurance, he didn’t die until after he had uttered the words, “Joy to you, we’ve won,” thereby informing the anxious Athenians that Sparta had defeated the invading Persians.
If Pheidippides’ accomplishment is worthy of celebration, so too are the therapeutic effects that we attribute to exogenous messenger RNAs, or mRNA-based drugs. These molecular messengers face their own marathon, the formidable expanse between in vitro transcription and in vivo delivery and translation. To help mRNA-based drugs traverse this ground and retain biological activity, drug developers are willing to play the role of trainer, identifying weaknesses and preparing therapeutic candidates to withstand the physical and chemical rigors of manufacturing workflows, routes of administration, and degradative metabolic pathways.
Drug developers are even willing to encapsulate mRNA drugs in nanoparticle delivery systems, the equivalent of giving a marathon runner a lift. No, this is not cheating. Surely, if Pheidippides had been offered a chariot ride, he would have accepted it, and the Athenians, full of rejoicing, would have approved.
Reinventing mRNA
The idea is rather simple: Construct and deliver an mRNA sequence to the cell and allow native machinery to crank out the encoded protein. The execution of this idea may be a little more complicated. Indeed, it may entail the reinvention of mRNA. For example, it may call for searches of mRNA databases to guide the engineering of mRNA that structurally resembles natural RNA. And that’s just a start.
The mRNA will also have to be modified for stability and packed into a suitable delivery vehicle. Only then will the mRNA have a chance of expressing a native protein or, possibly, a novel protein in the cytoplasm. By subjecting itself to the cell’s protein-expression machinery, the mRNA may serve as a template for an anticancer therapeutic or an immunostimulatory antigen.
Despite the difficulty of reinventing mRNA, drug developers remain optimistic. “It’s a powerful molecule with incredibly broad therapeutic potential,” declares Michael Heartlein, Ph.D., CSO, Translate Bio. Dr. Heartlein’s optimism, it should be emphasized, is validated by experience.
“Ten years ago, when we started our work, handling and manufacturing of large quantities of RNA meant ending up with micrograms or optimally milligrams,” he recalls. “Getting to the kilogram scale was a huge challenge. That was the first hurdle we cleared. We can now produce single batches of up to 100 g, and we have the processes in place to scale up to the kilograms necessary for commercialization of mRNA products.”
Another hurdle, Dr. Heartlein continues, is mRNA’s fragility: “The molecule breaks down easily and is susceptible to nucleases, hydrolysis, shear forces, etc.” This hurdle, too, is being cleared. “With creativity, collaborations, money, and a little luck,” asserts Dr. Heartlein, “we now have engineered ways to stabilize mRNA.”
Yet another hurdle that needs to be cleared is inefficient in vivo delivery. Exogenous mRNA must penetrate cells’ lipid membranes to reach the cytoplasm where it will be translated into functional protein. To clear this hurdle, Dr. Heartlein reports, Translate Bio developed special nanoparticle delivery vehicles. “These are less than 100 nanometers in size,” he notes, “and they differ in their formulations for different target tissues.” Each formulation reflects an effort to maximize uptake, release, and engagement of ribosomes.
Translate Bio’s work, Dr. Heartlein declares, has culminated in the company’s mRNA therapeutic (MRT) platform, which is designed to impact an array of diseases and target tissues using multiple routes of administration. For example, the company’s MRT5005 delivers an mRNA encoding a fully functional cystic fibrosis conductance regulator to the lung. It is currently in Phase I/II trials.
According to Dr. Heartlein, mRNA-based drugs hold great promise to expand the domain of “druggable” targets. “We feel that mRNA therapeutics are on the forefront of future therapies,” he concludes. “Today, we are making rapid headway in the development of first-generation approaches with the potential to transform treatment for a wide variety of diseases. In the future, with incremental advances and fundamental new scientific discoveries, we will see exciting next-generation therapeutics emerge.”
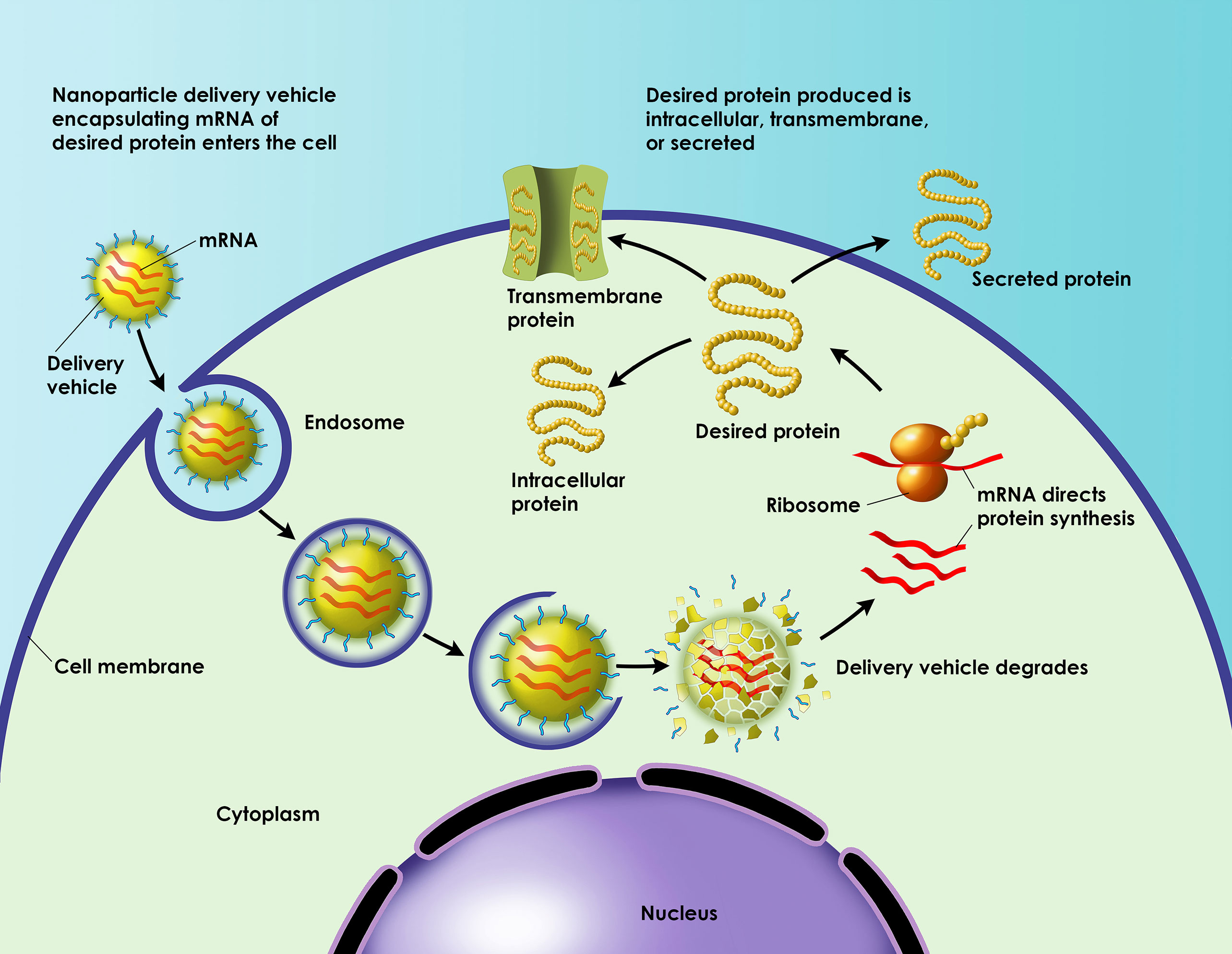
Platform Yields a New mRNA Vaccine
A new era in vaccinology is emerging now that conventional vaccines are being joined by mRNA vaccines. The newcomers offer high potency; that is, they can elicit an immune response even if they are present in minimal amounts. Also, they can be developed quickly and manufactured inexpensively.
Although early mRNA vaccines were faulted for their instability and inefficient delivery, these problems are being solved. New platforms are emerging that have the potential to produce clinic-ready prophylactic and therapeutic vaccinations.
Recently, Moderna Therapeutics utilized its mRNA platform to develop a vaccination strategy against cytomegalovirus (CMV). Currently, there are no approved vaccines for CMV, which is the leading cause of disabling infections in newborns. It is also the most frequent cause of viral disease in transplant recipients. CMV infections in transplant recipients often cause transplant failure.
To combat CMV, the company engineered and produced an mRNA vaccine candidate called mRNA-1647. It combines six mRNAs that encode six viral proteins: one protein is herpesvirus glycoprotein B (gB), a CMV antigen, and the other five proteins are units of the CMV glycoprotein H (gH) pentamer complex. Moderna encapsulated mRNA-1647 in a nanoparticle delivery system and assessed its performance in mice and nonhuman primate models.
“Past attempts at a vaccine for CMV were unable to include both of the important antigens required to block CMV infection, gB and the pentamer,” notes Michael Watson, MBChB, senior vice president of vaccine partnerships & health impact at Moderna. “The five-protein pentamer antigen is so complex, prior attempts could produce only the single protein gB antigen, but this left the cells that are unlocked by the pentamer unprotected.
“The main problem was trying to make such a complex protein outside of the body in a way that could be used for large-scale production. However, using our mRNA vaccine approach to make the protein in the body, just as the virus does, we have seen preclinical evidence that an mRNA vaccine can produce both the gB and pentameter proteins.
“In animal models, we’ve observed that the combination of the pentamer and glycoprotein B is highly immunogenic, raising high levels of neutralizing antibodies and strong T-cell responses. Our vaccine candidate for CMV, mRNA-1647, is now in Phase I trials, and we look forward to the results.”
Dr. Watson suggests that optimism is warranted by what he characterizes as Moderna’s success in clearing some major scientific hurdles in the production of mRNA-based vaccines and therapeutics. “We believe,” he says, “that mRNA medicines have tremendous potential for preventing and treating diseases that have eluded traditional drug development strategies.”
High-Throughput Analysis
Producers of mRNA-based therapeutics must assess product quality and quantity, and they must do so accurately and efficiently. “RNA therapeutics companies are looking more for high-throughput screening solutions that ensure reproducibility of RNA analyses, particularly those working with synthetic RNA fragments,” reports Denise Warzak, applications specialist, Advanced Analytical Technologies, Inc. (AATI), a part of Agilent Technologies. “For example, mRNA or RNA fragments are being synthesized for applications that include diagnostics and treatments for cancer and other diseases.”
Traditional methods for RNA analysis, such as agarose gel electrophoresis, can be inaccurate. Also, they tend to be limited to low-throughput processing. In contrast, contemporary methods, such as capillary electrophoresis (CE), facilitate high-throughput processing.
“Companies have strict requirements for digital readouts that check reproducibility, for example, as on hundreds of in vitro transcribed RNAs,” notes Steve Siembieda, AATI’s vice president of commercialization and marketing. “To drive efficiency and reduce costs, companies turn to automation for walkaway overnight runs that essentially extend the workday.”
AATI maintains a stable of automated CE systems that includes the Fragment Analyzer (the workhorse), the Fragment Analyzer Infinity (the adaptable endurance horse), and the FEMTO Pulse (the ultra-sensitive thoroughbred). These systems differ with respect to throughput, robotic systems interoperability, and sensitivity of detection. System-specific reagent kits are available from the company.
The more therapeutically minded users of AATI’s automated CE systems should maintain a broad outlook, suggests Siembieda. “There is a fascination with using single-cell transcriptome analysis to compare healthy and diseased cells,” he points out. “Ultimately, making personalized medicine a reality will likely require software that can merge information about the transcriptome with information from the genome, the metabolome, and other ‘omes’ that play into disease.”
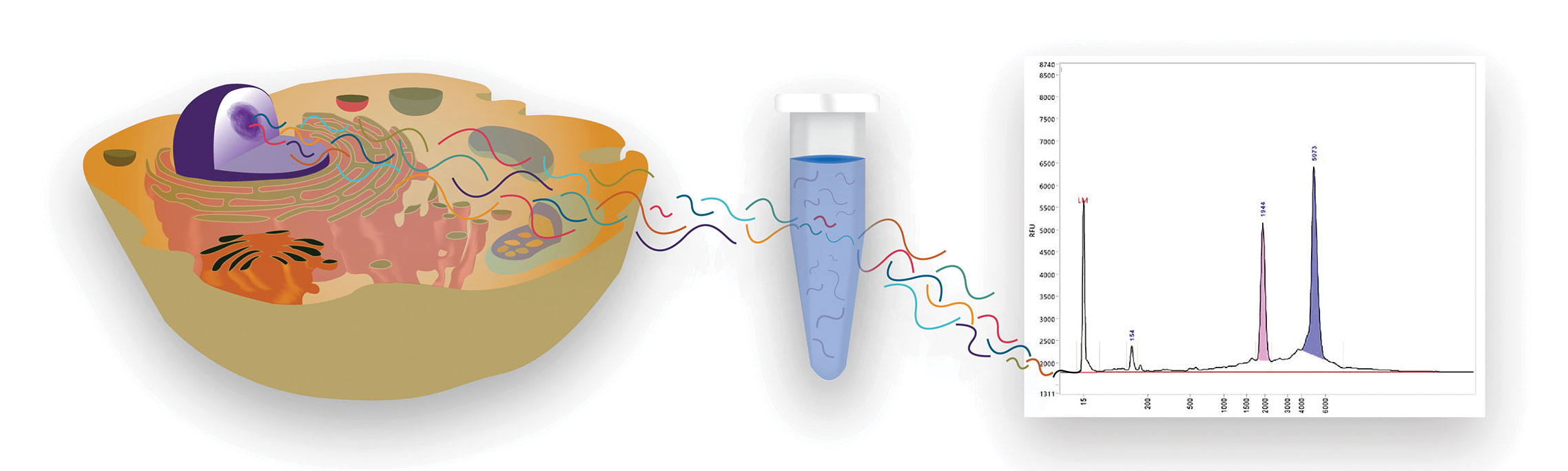
In Vitro Synthesis of mRNA
Many of the technical problems that arise during the development of RNA-based therapeutics can be solved if investigators use products from reliable vendors. One such vendor, Thermo Fisher Scientific, provides several tools that enable DNA and RNA therapeutic development and manufacturing workflows. For example, the company’s mMessage mMachine® SP6 transcription kit is designed for the in vitro synthesis of large amounts of capped RNA.
Capped RNA can mimic most eukaryotic mRNAs found in vivo because it incorporates a 7-methyl guanosine cap structure at the 5′ end. This structure is acquired through an ultra-high-yield transcription reaction. In the kit’s simplified reaction format, all four ribonucleotides and a cap analog are mixed in a single solution.
Special Needs
“Our company offers a variety of cell-based assays that allow researchers to analyze a drug’s impact on various pathways within a cell,” says Jennifer Loeb, global product manager, Promega. “Additionally, we offer several nucleic acid purification, quantitation, and amplification reagents to study transcriptomic and gene expression changes a drug may have. Finally, we also offer a variety of products which allow for RNA to be synthesized for use in in vitro and in vivo translation (for example, mRNA vaccine production).”
Investigators pursuing RNA therapeutics often have special RNA product needs. “Researchers developing RNA therapeutics are analyzing many potential drugs and keeping a fast pace,” Loeb notes. “This kind of work requires assays and methods that can be used in a high-throughput capacity. Investigators developing small interfering RNA or microRNA therapeutics require more complex and specialized drug delivery reagents compared to existing research small interfering RNA and microRNA delivery reagents.”
Although Promega has traditionally been viewed as a tools and reagents provider to companies working in this space, the company also has a team of scientists that works closely with companies to develop custom solutions. “We can help scientists as they move from discovery through production and quality testing,” Loeb asserts.
Keeping ahead of this fast-moving field is no easy matter. “RNA analysis continues to be a hot area of basic research and therapeutic development,” Loeb points out. “We monitor peer-reviewed literature, attend conferences, and talk to researchers to learn about the experiments or data that are causing difficulties.” For Promega, such difficulties represent opportunities. Addressing any one difficulty may prompt Promega to devise a custom solution, if that difficulty affects just one researcher, or a general solution, if that difficulty happens to affect multiple researchers.
The RNA therapeutics revolution has begun. Indeed, because it targets nucleic acids instead of proteins, the range of druggable targets expands well beyond that possible with small molecules or large biologics. Many in the field feel that new vaccines, cures for genetic diseases, and cancer treatments are all accomplishable goals.
Multiplexing Expands Scope of RNA Analysis
Sensitive, high-throughput techniques that enable the analysis of RNA in individual cells have been transformative for the fields of oncology, immunology, genomics, and developmental biology—revealing heterogeneity in populations of cells and identifying rare cell types for researchers to better understand biological processes. Analyzing several samples in one workflow via multiplexing, which is relatively new to the field of single-cell transcriptomics and multiomics, can take these capabilities to the next level, according to Nikhil Rao, Ph.D., global product marketing manager, BD.
“Multiplexing is an integral part of the evolution of RNA analysis,” asserts Dr. Rao. By incorporating multiplexing, researchers may overcome many of the technical hurdles that have limited RNA analysis to single cells. Researchers may also increase study throughput and collect more reliable cell data.
“Many of the commercially available platforms for RNA single-cell analysis considered to be high-throughput systems can process only one sample at a time, limiting a researcher’s ability to add controls,” he notes. Multiplexing can increase sample throughput, minimize batch effects, and identify multiplets to provide a more controlled and economical experiment.
“Sample multiplexing allows faster analysis and more reliable experimental insight. By adding more samples on a single run, you can reduce single-cell library preparation reagent cost, save time by executing fewer experiments, and detect intersample multiplets,” Dr. Rao explains. “In some cases, multiplets are not easily identifiable, and they can be misinterpreted as biologically meaningful.
“The addition of sample-specific antibody tags for individual cells, like those in the BD Single-Cell Multiplexing Kit, allows researchers to combine and load up to 12 samples while improving data quality by maintaining a low rate of unidentified multiplets in data analysis.”
Evaluating How RNA Analysis Is Affected by Modified RNA Bases
Jennifer L. Ong, Ph.D.
A rapidly growing field of study is focused on modifications to the standard RNA bases. These modifications, which are chemically diverse and present in the mRNA of various organisms, can alter gene expression or mRNA stability. They are also involved in various cellular processes and diseases. Accordingly, there is great interest in detecting RNA modifications, but to date there is only limited knowledge about how these modifications affect the fidelity and performance of biotechnology enzymes used in RNA analysis.
To investigate these enzyme effects, we designed a fidelity assay based on Pacific Biosciences’ single-molecule real-time (SMRT) sequencing, which is capable of sequencing individual DNA molecules with very high accuracy. We measured the combined error rates for T7 RNA polymerase and Protoscript® II reverse transcriptase, with both standard RNA and modified RNA, including RNA containing biologically significant RNA base modifications.
These modifications include N6-methyladenosine (m6A), pseudouridine (Ψ), 5-methylcytidine (m5C), 5-methyluridine (m5U), and 5-hydroxymethyluridine (hm5U). m6A, Ψ, and hm5U increased first-strand error rates, whereas m5C and m5U did not change the combined fidelity of T7 RNA polymerase and Protoscript II.
In addition to measuring error rates, we also identified modification-induced substitution errors. Our results are an important step toward understanding the role of modified RNA in biotechnology. Our findings also have implications for synthesizing modified RNA for therapeutics and detecting RNA modifications by next-generation sequencing.
Jennifer Ong, Ph.D., is scientific director, nucleic acid replication, New England Biolabs.
Ensure RNA Quality during Cancer Transcriptome Profiling
Researchers interested in transcriptome profiling need high-quality RNA. This can be especially difficult to obtain for cancer researchers who need to study RNA expression at a cell- or location-specific level because of the heterogeneity of tumors. Only a combination of precise sampling and effective QC allows cancer researchers to capture quality transcriptomic data.
Laser capture microdissection (LCM) can isolate individual tumor cells, but it requires that tissue be preserved using methods, such as freezing and fixation, that can degrade RNA. Oncology researchers should monitor RNA quality during LCM to ensure transcripts are of sufficient quality to produce reliable results in RT-PCR, microarray analysis, and NGS. By determining RNA quality before performing further downstream analysis, researchers save time and costs they would otherwise waste on poor-quality samples.
Traditional RNA analysis methods (such as RNA agarose gel electrophoresis) lack the throughput, sensitivity, and accuracy required to assess RNA quality, according to Steve Siembieda, vice president of commercialization, Advanced Analytical Technologies, Inc. (AATI), a part of Agilent Technologies. They can also be tedious and time consuming, he adds.
“For RNA quality analysis, AATI designed the Fragment Analyzer™ Automated CE System, which uses parallel capillary electrophoresis to deliver reliable RNA assessment in under one hour for samples with concentrations as low as 50 pg/µL,” says Siembieda. “Up to 288 samples can be processed without intervention, ideal for high-throughput genomics labs.”
Specifically, the Fragment Analyzer provides the RNA quality number (RQN), which indicates the extent of RNA degradation on a 10-point scale, where 1 represents totally degraded RNA, explains Siembieda. “The RQN,” he continues, “accounts for several parameters such as the 28S:18S ribosomal RNA ratio, wherein a lower number indicates an increased concentration of shorter or degraded RNA, and the 5S region, where small RNA separates.”