![Loss of TET function contributes to skewing blood stem cells in favor of forming myeloid cells over other blood cell types by regulating the expression of lineage-specific genes. [Dr. Myunggon Ko, Ulsan National Institute of Science and Technology]](https://genengnews.com/wp-content/uploads/2018/08/Dec10_2015_UlsanNationalInstScienceTech_TETFunction1423481231-1.jpg)
Loss of TET function contributes to skewing blood stem cells in favor of forming myeloid cells over other blood cell types by regulating the expression of lineage-specific genes. [Dr. Myunggon Ko, Ulsan National Institute of Science and Technology]
Scientists at the La Jolla Institute for Allergy and Immunology say they have demonstrated that TET (ten-eleven translocation) proteins collectively constitute a major class of tumor suppressors and are required to maintain genome stability. Their study (“Acute loss of TET function results in aggressive myeloid cancer”), published in Nature Communications, shows that mice lacking both Tet2 and Tet3 rapidly develop an aggressive form of myeloid leukemia. The researchers believe that a better understanding of genome instability may open new avenues for treating cancer.
“We removed Tet2 and Tet3 by injecting the mice with chemicals that acutely deleted their genes, after which we could see signs of cancer in one week and a full-blown transmissible cancer in four weeks,” says senior author Anjana Rao, Ph.D., a professor in the institute's division of signaling and gene expression who led the study. “This proves that TET-loss of function directly caused the malignancy.”
The gene coding for Tet2 is frequently mutated in patients with myeloid malignancies and, even without Tet2 mutations, many human myeloid cancers display low TET function as Dr. Rao and her team had previously demonstrated. “The same is true for many other types of cancers,” says the study's co-author Myunggon Ko, Ph.D., formerly a postdoctoral researcher in the Rao lab and now an assistant professor at the Ulsan National Institute of Science and Technology in Ulsan, Korea. “Thus our data imply that TET function is likely to be important for preventing not only myeloid cancers, but also potentially other types of cancers.”
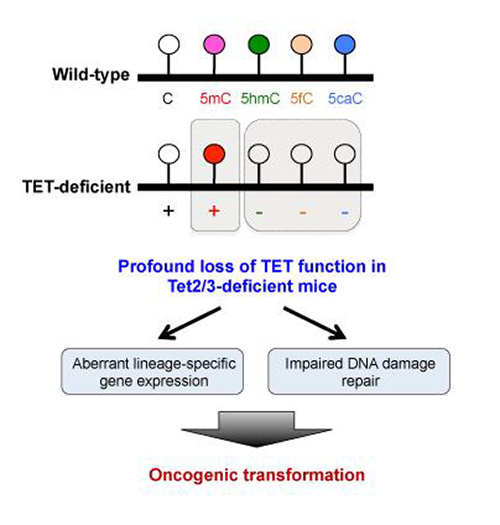
Members of the TET family of enzymes help rewrite the epigenome and help determine gene activity without changing the genetic code. Specifically, TET enzymes convert a modified form of cytosine, 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC) and other forms of oxidized methylcytosine. Together these oxidized forms of 5mC facilitate DNA demethylation but are also important in their own right.
“Our findings indicate that loss of TET function contributes to skewing blood stem cells in favor of forming myeloid cells over other blood cell types by regulating the expression of lineage-specific genes,” says Dr. Ko. “Surprisingly, although cytosine methylation has long been thought to inhibit the activity of genes, there was no clear correlation between DNA methylation and gene expression. They were independently controlled and we may have to change our view about the relation between DNA methylation and gene expression.”
“One possibility is that the precipitating factor is loss of oxidized methylcytosines rather than changes in DNA methylation per se,” adds Dr. Rao.
Deleting just one of the three members of the TET family (Tet1, Tet2 and Tet3), although insufficient to induce full-blown leukemia in a model system, leads to the expansion of the stem cells that give rise to all blood cells. This process is exacerbated in stem cells with more severely impaired TET function, such as that seen in this study in mice lacking both Tet2 and Tet3. The capacity for self-renewal is enhanced and the cells preferentially differentiate into cells belonging to the myeloid lineage, such as macrophages and neutrophils.
The researchers also observed a strong impairment of DNA damage repair, which suggests an important role for TET proteins in maintaining genome stability. “Genome instability is a defining characteristic of most cancers and a direct result of defective DNA damage repair,” says Dr. Ko. “What we don't know yet is how TET loss of function leads to impaired DNA damage repair.”