If a genetic variant is the origin and a disease the destination, the biochemical path between them may appear on a map of a sprawling genetic/metabolomic network. But that’s just the simplest imaginable use of such a map. Rather than think of a genetic/metabolomic map as a way to trace a single path, as though one were using a transit map to trace an individual’s commute, one might think globally, in terms analogous to those used by traffic managers. For example, what snarls might arise if multiple lines and transfer nodes were to become overloaded?
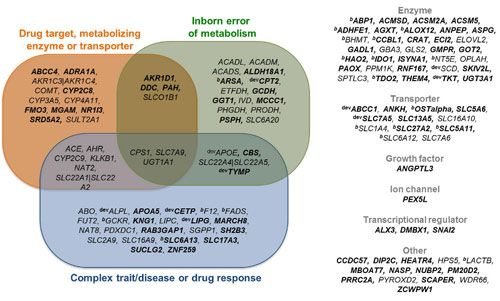
This approach to analyzing the genetic influences on metabolism, and metabolic diseases, is becoming a reality thanks to projects such as the one recently completed by researchers based at the Wellcome Trust Sanger Institute. These researchers, led by Nicole Soranzo, Ph.D., have compiled an atlas of genetic associations with metabolism that has linked 145 genetic regions with more than 400 metabolites in human blood. This new compendium of associations between genetic regions and metabolite levels provides a powerful tool to identify genes that could be used in drug and diagnostic tests for a wide range of metabolic disorders.
The research team’s results were presented May 11 in Nature Genetics, in an article entitled “An atlas of genetic influences on human blood metabolites.” In this article, the authors wrote: “Our observations suggest widespread genetic control over a large range of different pathways and functions and support the notion of human metabolism as a complex continuum governed by genetic effects of variable intensity, complex regulatory influences, and nongenetic effects. Our results advance knowledge in a number of areas of biomedical and pharmacogenetic interest, generating nearly 100 new hypotheses of SNP-metabolite and disease correlates and identifying a large catalog of new potential biomarkers as well as associations to drug targets, transporters, and metabolic enzymes.”
“The sheer wealth of biological information we have uncovered is extraordinary” said Dr. Soranzo. “It’s exciting to think that researchers can now take this freely available information forward to better understand the molecular underpinnings of a vast range of metabolic associations.”
This last comment reflects the ready availability of the data and results generated by the Wellcome-led study. As indicated in the Nature Genetics paper, the researchers “developed a database and web-based resources for data mining and results visualization.”
“We developed an open-access database that allows researchers to easily search through the findings, to understand genetic variants associated with metabolism one metabolite at a time and in the context of the complete metabolic network,” added Gabi Kastenmüller, Ph.D., co-senior author from the Helmholtz Center Munich, Germany. “This database will facilitate drug discovery for metabolic disorders and also help researchers to understand the biology behind disease.”
The researchers found 90 new genetic associations, trebling the figure of known genetic associations with metabolites. In many of the cases where metabolites were known, the team were able to link the molecule to gene function. They mapped genes to their likely substrates or products and linked these to a number of conditions, including hypertension, cardiovascular disease, and diabetes.
Other associations suggest tantalizing possibilities for further study. For instance, a number of the genetic associations identified involved aromatic acids, such as tryptophan, which are important for brain function. While this study did not measure association of metabolites in the brain, these genetic findings open new avenues to assess potential genetic influences on brain function and responses to drugs that affect brain function, such as antidepressants.
“This work provides an important new window into the genetic variation underlying human metabolism,” commented Eric Fauman, Ph.D., study co-author and associate research fellow from Pfizer. “Through targeted precision medicine and by linking human disease genes to in vivo biological markers, we hope to enhance our ability to deliver impactful new medicines for patients across a variety of disorders.”