Last week, Jennifer Doudna and Emmanuelle Charpentier, winners of the Nobel Prize for Chemistry in 2020 for developing CRISPR gene editing technology, belatedly traveled to Stockholm to enjoy the pageantry and ceremonies that were postponed two years ago because of the COVID pandemic. But as scientists celebrate the revolutionary development of CRISPR-Cas9 genome editing just ten years ago, they continue to develop new technologies for manipulating DNA.
CRISPR is a superb programmable scissor but has certain drawbacks. After making a double-stranded break in DNA, it is up to the cell’s DNA repair pathways to rescue and effectively fix that broken DNA. Because the repair pathways evolved to fix broken DNA quickly, it’s hard to use these nucleases to introduce a new sequence or gene into the genome, install a specific type of edit, or change one base to another or many bases all at once. For years, genome engineers have been trying different tricks to increase the efficiencies of these repair pathways to achieve specific goals without any clear breakthroughs.

A few years ago, Jacob Rubens, senior principal at Flagship Pioneering, and his colleagues asked whether nature, instead of creating mechanisms for breaking DNA, had evolved schemes for writing DNA. “It didn’t take long before we came to mobile genetic elements—sequences of DNA that exist for the sole purpose of copying and pasting themselves from one location in the genome to another,” Rubens told GEN Edge. “We didn’t know that these are the most abundant genes in nature!”
It turns out that there’s more real estate in genomes that codes for this mobile genetic element machinery than any other protein class—transporters, metabolic enzymes, etc. “It’s not surprising considering these elements are just copying and pasting themselves everywhere,” said Rubens. “But Mother Nature has been evolving these systems for billions of years, and we were surprised to discover that almost no one had thought about how they could be adapted as tools for genome engineering, even though their sole purpose is to write DNA.”
Rubens and his team opened a thousand-page textbook called Mobile DNA III that was edited by Nancy Craig, one of the leading researchers in the field. It contains dozens of chapters on different types of mobile genetic elements. Yet, Rubens said that almost all the relevant work within the space of biotechnology could fit into just a couple of those small chapters. They focused on a single type of mobile genetic element known as a DNA transposon, which has some applicability in biotechnology but had been somewhat limited.
Rubens and his team found that there are many other types of biochemistries that could become biotechnologies but that had not been studied. This gave them some ideas about different types of mobile genetic elements suitable for genome engineering because of their features and abilities. And that’s how Tessera Therapeutics was born. The company name has two origins: “tessares” is Greek for the number 4, as in four nucleotides in the genome. And a tessera is a tile in a mosaic. So, the name for the company comes from a view that every nucleotide is like one tile in the mosaic of life.
“We put a couple of million dollars into seeding the company in 2018 and hired our first scientists, finding some fantastic people with experience in gene editing companies like Editas, Intellia, and Beam to come aboard,” said Rubens. For example, Tessera was able to recruit the likes of Senior Vice President Cecilia Cotta-Ramusino from Editas and Chief Medical and Development Officer David Davidson from bluebird bio. Chief Scientific Officer (CSO) Michael Holmes, who was one of the star scientists at Sangamo and developed the concept of genome editing with zinc finger nucleases with Fyodor Urnov, joined from Ambys Medicines. Tessera’s Scientific Advisory Board is stacked, with Board Director Melissa Moore (previous Moderna CSO), gene transfer pioneer Luigi Naldini, Innovative Genomics Institute (IGI) co-director and co-founder of KSQ therapeutics and Maze therapeutics Jonathan Weissman, and the infamous George Church, to name a few.
“We began to conduct some experiments and realized there was a lot of potential here and what it would take to truly build a breakthrough company. We realized that what it was truly going to take to build the biggest version of this company was not just trying different types of mobile genetic elements but systematically exploring the types of elements that had interesting biochemistries for the functions that we wanted.”
Harnessing mobile genomic elements
Over the next few years, the team at Tessera built the capability to make thousands of mobile element sequences per month in DNA and RNA formats, put them on human cells, and measure tens of thousands of different changes in the genome monthly in different cell types to figure out which of these systems work for their purposes.
Tessera has invested heavily in high-throughput DNA and RNA manufacturing to do massive amounts of high-throughput screening. Rubens said that it is routine for Tessera to make thousands of DNA plasmids each month, which are typically turned into hundreds of different RNAs. These RNAs are then put into cells using automated cell-handling machinery for transfections and cell passages.
Next, they use machines to extract the nucleic acids from the cells and test for three key properties that are important for a good genome engineering technology: efficiency (the percent of alleles or cells in the population that are effectively edited); specificity (the location of those edits in the genome); and fidelity (does the appropriate outcome occur at the intended site?).
There are different automated pipelines for each run at Tessera. The most important one currently is the assay that measures efficiency, called amplicon sequencing—the same type of sequencing reaction for figuring out the SARS-CoV-2 variants in patients. With a targeted polymerase chain reaction and sequencing, it is possible to survey the genome to see where changes are made by the machinery that writes genes. Very soon, Tessera will have end-to-end automation to do 20,000 sequencing reactions per month.
Rubens says that Tessera has also built the data science infrastructure to ingest data from a variety of experiments and to leverage the differences in the performance of these systems to inform how they change the compositions that matter, whether those are the amino-acid sequences of the gene writers or the RNA sequence or DNA sequence of the templates, to make them work more effectively towards those parameters that they measure.
RNA writing medicines
Of course, Tessera is not merely a technology company. It plans on creating medicines, starting with a few particular patient populations – and several oncology applications (both yet to be disclosed).
Tessera’s technology, called RNA gene writers, is based upon a type of mobile element called a non-LTR retrotransposon. These use a mechanism to write genes into the genome called target primed reverse transcription, which Rubens thinks will be a hot topic over the next few years. The method relies upon four key steps: binding RNA, binding DNA, nicking DNA, and then priming reverse transcription (Fig. 1).
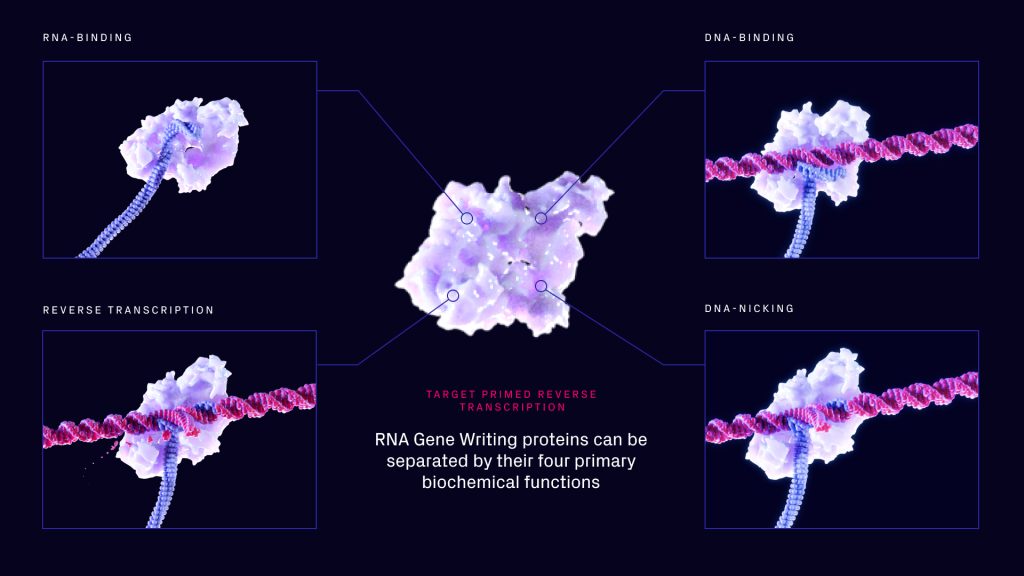
According to Rubens, Tessera saw right away that the mechanism had some interesting aspects. First, it’s very programmable in that it’s possible to target genome sites for editing or integration. Second, it only makes use of an RNA template (Fig. 2). This makes it possible to make systems that can write whole genes into the genome by sending RNA to the cell. This is done by sending an mRNA that codes for the gene writer and a template RNA. The gene writer then grabs the template RNA and writes it into the genome.
“That opens up amazing possibilities because it permits us to bring together the very best of mRNA and lipid nanoparticles (LNPs) with the very best of gene therapies—the manufacturability of mRNAs and LNPs, the scalability, and the re-dosability with the longevity of viral vector gene therapies,” said Rubens.
There are two major potential clinical applications for Tessera’s technology:
- Writing—adding entire genes to the genome with their promoters and other parts necessary for expression; and
- Rewriting—to make edits to the genome (small substitutions, insertions, and deletions).
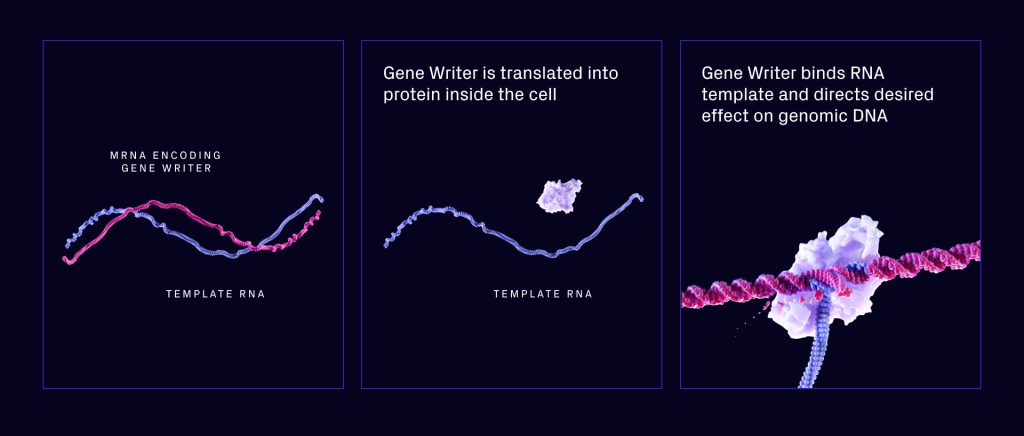
In the short term, Rubens said one area of focus is writing chimeric antigen receptors (CARs), which are cells that reprogram immune cells to target cancer.
“The vast majority of people developing CAR-T therapies take cells out of a patient’s body and treat the patient with some toxic drugs that clear out their endogenous T-cells. Then they engineer the patient’s cells with a lentiviral vector and reinfuse that into patients,” Rubens explained. “By doing CAR-T engineering with just RNA, we can not only do this faster, but we can also do it in vivo with mRNA and lipid nanoparticles.”
Regarding their rewriting technology, Tessera aims to make small genomic substitutions. “Right now, we’re primarily focused on two areas: liver-targeted editing applications, such as phenylketonuria (PKU), where we’ve shown really exciting progress by editing 40% of the [mutant] alleles in a liver with clear curing of PKU phenotype in the ENU2 mouse model of this disease, said Rubens.
The other example is sickle-cell disease (SCD). Rubens continued: “We’re the only ones (that we’re aware of) that is developing a therapy for sickle cell disease, where we can just deliver RNA into a cell and correct the allele that causes the disease to wild-type sequence. Not only that, but because it’s all RNA and in LNPs, we’ve made huge strides in delivering it to hematopoietic stem cells inside the body. So, like the CAR-T example, we no longer have to take the cells out of a patient’s body, engineer them, and return them.”
This is important because the biggest burden of SCD—in the United States at least—falls on patients who can’t take weeks off work to go through the therapy required to receive a cure. Most SCD patients live in sub-Saharan Africa. “If we want to be able to help them, we need to be producing drugs at the same cost of goods at which we’re producing the current virus vaccines—the price of a fancy T-shirt—and that’s uniquely enabled today by our RNA writing technology,” said Rubens.
In recent conference presentations, Tessera scientists have shared some primary proof-of-concept data from animals for liver rewriting and T-cell therapies. “We are engineering T cells outside the body and delivering them to monkeys, as well as delivering to T cells and hematopoietic stem cells inside of monkeys,” said Rubens. “So, it is very real, and we’re going to hopefully share more soon.”
DNA writing medicines
When launched in 2018, the team at Tessera also began working on serine integrases and discovered 75,000 serine integrases. Unlike the approach that the groups of (twinPE) have taken, Tessera has been searching for serine integrases that target native sites on the human genome (similar to a recently published approach from Patrick Hsu’s group at the Arc Institute). In a screen of some 750 enzymes, Tessera found a handful that were very efficient and quite specific at integrating genes into the genome without having to do the first step of the PASTE process using prime editing to write a landing pad.
Tessera has developed a simpler version of these systems. With a related class of DNA gene writers, the company has shown that the combination of DNA gene writers and adeno-associated viruses (AAVs) has potential, at least in mice and non-human primates, to make gene expression more permanent by writing it into the genomes of the animals.
By writing an AAV into the genome of dividing cells such as the liver, Tessera can make the AAV express permanently. This is critical for certain applications, such as treating newborn patients with AAV gene therapies. Because the liver divides over the first 18 years of the patient’s life, they typically aren’t candidates for these gene therapies today.
“We’ve shown in mice that we can make 40% of their hepatocytes express the payload we delivered over the lifetime of these animals, compared to just 2–3% in the mice that receive the control AAV,” said Rubens. “And we’ve demonstrated something similar in non-human primates now, but we haven’t really shared that data publicly yet.”
Tessera has also shown an ability to integrate AAVs into the genomes of adult mice. “When those AAVs integrate into the genome, we’ve shown that they express at an 85-fold higher level,” said Rubens. “That means we could reduce the AAV dose by 85-fold to achieve the same level of therapeutic protein expression. This would likely solve a lot of the problems that AAV has in facing dose-limiting toxicities that we hear about all the time in the clinic, as well as the manufacturability issues. We’ve also shown that this property is translatable from the mice to the non-human primates.”
A Marathon long sprint
What started in a small basement in Kendall Square with a few former alums of these genetic medicine companies is 275 people today with close to 90,000 square feet of space at a facility in Somerville, Massachusetts, where Flagship has a few different companies co-located.
“We expect to have proof of concept that our technologies work in non-human primates shortly, and just a couple of years after that, we expect to put our first drugs in the clinic,” said Rubens. “It’s been a quick sprint from the fundamental idea, the perfect concept, to starting to make compositions that matter, that someone might use to help people, which has been fascinating to watch.”