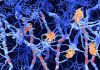
Several hundred Cas9 enzymes (red dots) searching the nucleus of a live mammalian cell for a particular DNA sequence. They become white when they bind briefly before moving on. The lines show the paths these enzymes have taken, color coded according to time. The Cas9 tracks show that the enzymes search by diffusing through the nucleus, and that off-target binding events are predominately short lived. Motion is slowed to half normal speed. [Spencer Knight video, UC Berkeley]
From the co-discoverer of the CRISPR-Cas9 system and her colleagues at UC Berkeley comes new data that should instill a greater level of confidence that the genome editing tool won't inadvertently excise off-target DNA.
The two new reports from the Berkeley investigators show in great detail how the Cas9 protein searches through billions of base pairs in a cell to find the right DNA sequence, and how Cas9 determines whether to bind or bind and cut, thereby initiating gene editing. From the collected data, the researchers were able to surmise that Cas9 appears to have at least three ways of checking to make sure it finds the right target DNA before it takes the irrevocable step of making a cut.
“CRISPR-Cas9 has evolved for accurate DNA targeting, and we now understand the molecular basis for its seek-and-cleave activity, which helps limit off-target DNA editing,” explained senior author of both articles and co-discover of the CRISPR-Cas9 system Jennifer Doudna, Ph.D., a Howard Hughes Medical Institute investigator and professor of molecular and cell biology and of chemistry at UC Berkeley.
The findings from this study were published recently in two papers: the first in Science through an article entitled “Dynamics of CRISPR-Cas9 genome interrogation in living cells” and the second in Nature in an article titled “Conformational control of DNA target cleavage by CRISPR–Cas9.”
In the Science article, the researchers tracked Cas9-RNA molecules within the nucleus of mammalian cells as they rapidly searched through the entire genome to find and bind to their prescribed target sequence.
“It's crazy that the Cas9 complex manages to scan the vast space of eukaryotic genomes,” remarked lead author of the Science paper and graduate student Spencer Knight. “There is a lot of off-target binding by Cas9, but we found that these interactions are very brief—from milliseconds to seconds—before Cas9 moves on.”
The Berkeley team estimated that a few thousand CRISPR-Cas9 complexes can scour the entire genome extremely rapidly to find one targeted stretch of DNA. Additionally, Cas9 must also recognize a short three-base-pair DNA sequence immediately following the primer sequence, or PAM, which occurs roughly 300 million times within the human genome.
“If Cas9 bound for tens of seconds or minutes at each off-target site, it would never, ever be able to find a target and cut in a timely manner,” Knight said.
In the Nature article, the investigators observed that once Cas9 binds to a region of DNA, it performs an additional check before two distal sections of the Cas9 protein complex come together, like the blades of a scissors, to precisely align the active sites that cut double-stranded DNA.
“We found that RNA-guided Cas9 can bind some off-target DNA sequences, which differ from the correct target by just a few mutations, very tightly. Surprisingly, though, the region of Cas9 that does the cutting is inhibited because of the imperfect match. But when the correctly matching DNA is located, Cas9 undergoes a large structural change that releases this inhibition and triggers DNA cutting,” noted the lead author of the Nature paper Samuel Sternberg, Ph.D., former graduate student in Dr. Doudna’s laboratory.
“We think that this structural change is the last checkpoint, or proofreading stage, of the DNA targeting reaction,” Dr. Sternberg continued. “First, Cas9 recognizes a short DNA segment next to the target—the PAM—then the target DNA is matched up with the guide RNA via Watson-Crick base-pairing. Finally, when a perfect match is identified, the last part of the protein swings into place to enable cutting and initiate genome editing.”
When asked about the impact of the current researcher, Knight responded that the data “suggests that you have more than one checkpoint to ensure correct Cas9 binding. There's not just sequence regulation, there is also temporal regulation: it has to engage with the DNA and park long enough that it can actually rearrange and cut.”
The discoveries from the Berkeley teams should allow for greater insight into the molecular mechanisms that lead to off-target events that can occur during genome editing applications—an critical area for researches to grasp an understanding of should they hope to use CRISPR-Cas9 in a wider clinical capacity.