Source: Mol Syst Biol. (2018) 14: e8242
Scientists from the European Molecular Biology Laboratory (EMBL) say they have adapted an existing technique to study the melting behavior of proteins so that it can be used for the study of bacteria. Antibiotic resistance is spreading worldwide so there is a strong need for new technologies to study bacteria, according to the researchers who published their study (“Thermal proteome profiling in bacteria: probing protein state in vivo”) in Molecular Systems Biology.
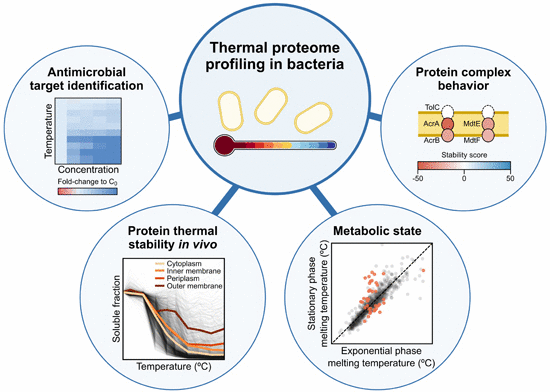
“We adapted thermal proteome profiling (TPP) to probe the thermostability of Escherichia coli proteins in vivo. E. coli had a more thermostable proteome than human cells, with protein thermostability depending on subcellular location—forming a high-to-low gradient from the cell surface to the cytoplasm. While subunits of protein complexes residing in one compartment melted similarly, protein complexes spanning compartments often had their subunits melting in a location-wise manner. Monitoring the E. coli meltome and proteome at different growth phases captured changes in metabolism. Cells lacking TolC, a component of multiple efflux pumps, exhibited major physiological changes, including differential thermostability and levels of its interaction partners, signaling cascades, and periplasmic quality control,” write the investigators.
“Finally, we combined in vitro and in vivo TPP to identify targets of known antimicrobial drugs and to map their downstream effects. In conclusion, we demonstrate that TPP can be used in bacteria to probe protein complex architecture, metabolic pathways, and intracellular drug target engagement.”
TPP enables scientists to compare the melting behavior of all proteins in a cell or organism before and after a perturbation, such as the administration of a drug. By adapting TPP to bacteria, it can now be used to study the activity and architecture of most proteins in a bacterial cell while it's alive, notes André Mateus, Ph.D., a postdoc working in the Savitski and Typas groups at EMBL.
While human bodies cease to function at temperatures above 42°C, E. coli bacteria still grow regularly up to 45°C. “We discovered that proteins in the middle of a bacterial cell are less tolerant to heat than those at the cell surface,” says Mikhail Savitski, Ph.D., team leader and head of proteomics core facility, EMBL. “Surprisingly, a protein's location is more predictive for its melting behavior than which other proteins it interacts with.”
With TPP, researchers can also investigate the effects of drugs on bacteria. Protein-drug interactions typically increase the proteins' heat tolerance, resulting in higher melting points. Therefore, comparing the heat tolerance of drug-treated and untreated bacterial cells helps to identify targets of antimicrobial drugs, but also to decipher how the bacterial cell succumbs to the drug or tries to bypass its action.
“In one particular case, we were able to elucidate a novel drug resistance mechanism,” says Dr. Mateus. “Cells use proteins to pump antibiotics out of the cell. After genetically removing one such efflux pump from their chromosome, bacteria became more sensitive to many drugs but curiously more resistant to one specific antibiotic called aztreonam. Using TPP, we found that this was due to dramatically reduced levels of a specific porin – a protein that acts as a pore – used by aztreonam to enter the cell.”
Compared to other techniques, TPP allows scientists to investigate the effects of perturbations on thousands of individual proteins in a short timeframe, explain the EMBL researchers. Most of the obtained insights – like the changes in the activity of proteins in vivo – would be impossible with other conventional techniques and for so many proteins simultaneously, showing TPP's potential to study bacteria in detail, according to the scientists.

![<em>E. coli</em> bacteria colored in heat gradient. [Aleksandra Krolik/EMBL] <em>E. coli</em> bacteria colored in heat gradient. [Aleksandra Krolik/EMBL]](https://www.genengnews.com/wp-content/uploads/2018/08/Jul9_2018_EMBL_EColi1952311591.jpg)
