Boston College chemists say they have developed a technology to precisely incorporate a range of useful non-canonical amino acids into proteins made in eukaryotes. The team’s goal was to create a new method to engineer and monitor protein functions as a way of expanding the scientific understanding of the processes that guide protein functions in our cells.
Their study (“Resurrecting the Bacterial Tyrosyl-tRNA Synthetase/tRNA Pair for Expanding the Genetic Code of Both E. coli and Eukaryotes”) appears in Cell Chemical Biology.
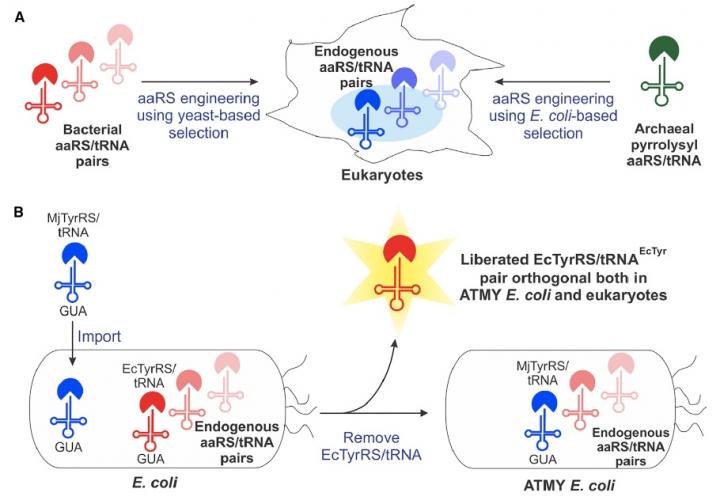
“The bacteria-derived tyrosyl-tRNA synthetase (TyrRS)/tRNA pair was first used for unnatural amino acid (Uaa) mutagenesis in eukaryotic cells over 15 years ago. It provides an ideal platform to genetically encode numerous useful Uaas in eukaryotes. However, this pair has been engineered to charge only a small collection of Uaas to date. Development of Uaa-selective variants of this pair has been limited by technical challenges associated with a yeast-based directed evolution platform, which is currently required to alter its substrate specificity,” write the investigators.
“Here we overcome this limitation by enabling its directed evolution in an engineered strain of E. coli (ATMY), where the endogenous TyrRS/tRNA pair has been functionally replaced with an archaeal counterpart. The facile E. coli-based selection system enabled rapid engineering of this pair to develop variants that selectively incorporate various Uaas, including p-boronophenylalanine, into proteins expressed in mammalian cells as well as in the ATMY strain of E. coli.”
According to Abhishek Chatterjee, Ph.D., assistant professor of chemistry, and lead on the project, the researchers were surprised by the facility of the new approach.
“Creating this novel E. coli strain required substituting its native aminoacyl-tRNA synthetase/tRNA pair with a counterpart from a different organism, which we anticipated would be very difficult,” he says. “But it turned out to be quite feasible. That opens up this complete technology.
“Thousands of proteins are encoded in the genome that make us who we are, but we know very little about that process. In human cells, there are roughly 20,000 protein-coding genes. What they are doing and how they are doing it remains difficult to study. One of the major problems is that if you want to know what they are doing, you have to spy on them. You need to attach a probe that can report back on what is going on.”
Introducing such probes has proven difficult, as the process often damages the target protein.
“The idea is that we can introduce a new building block into proteins that nature does not have—beyond the 20 canonical amino acids that nature uses,” Dr. Chatterjee says. “If we can do that, we have the ability to very specifically introduce a wide variety of non-natural functionalities into any site of virtually any protein.”
The immediate benefit would be to assist researchers who are still unraveling the mysteries of cell biology and protein function.
“You could create a protein with a non-canonical amino acid into any chosen site, load it with probes that are very tiny and give out an optical signal that tells where it is going,” continues Dr. Chatterjee. “It could allow you to manipulate how the protein is working. You could introduce limits, so whatever the protein is doing, it can't do anymore. And you could remove the probe by using an external signal such as light. This technology opens up numerous new ways one can start to probe and engineer protein function, which would be very challenging otherwise.”