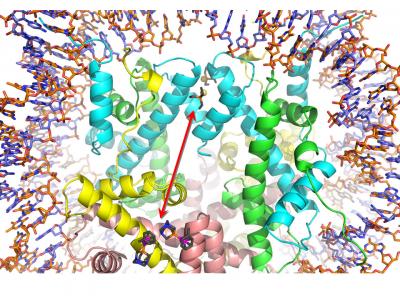
![Close-up of the histone protein region of a nucleosome core particle in which two different drugs (auranofin and RAPTA-T) are bound. [Paul Dyson/EPFL]](https://genengnews.com/wp-content/uploads/2018/08/Mar30_2017_PaulDysonEPFL_HistoneProtein3512016142-1.jpg)
Close-up of the histone protein region of a nucleosome core particle in which two different drugs (auranofin and RAPTA-T) are bound. [Paul Dyson/EPFL]
Two unrelated drugs, an anticancer agent and an antirheumatic, kill more cancer cells than either can kill alone. As a team, the drugs can take advantage of an allosteric, “action at a distance” mechanism. Essentially, the binding of one drug alters the activity of the other.
The anticancer drug, RAPTA-T, is a ruthenium-containing anticancer drug that disrupts both tumor growth and metastasis, while also reducing the side effects of chemotherapy due to its low toxicity. To achieve these effects, it forms adducts with the histone proteins that package DNA. These adducts disrupt the normal function of DNA and cause the cell to die.
The antirheumatic, auranofin (Ridaura®), is a gold-containing drug that is used to alleviate the symptoms of rheumatoid arthritis. Auranofin is much less prone than is RAPTA-T to form adducts with the histone proteins, unless the two drugs are used together.
Although the two drugs are used for different conditions, auranofin has recently been discovered to also act against cancer. The reason is that, often, drugs do not only bind a single site on a specific molecule, but can also bind and affect other, unrelated sites—either on the same molecule or on a different one. For example, a drug that is meant to bind and activate a receptor could also bind and block an enzyme. This off-site activity frequently gives rise to drug side effects, but separate drug-binding sites can also work together synergistically in a productive fashion.
The drug-drug synergy between RAPTA-T and auranofin was recently investigated by scientists based at École polytechnique fédérale de Lausanne (EPFL) and Nanyang Technological University (NTU). The labs of Paul Dyson, Ph.D., and Ursula Röthlisberger, Ph.D., at EPFL, together with the lab of Curtis Davey, Ph.D., at NTU, looked at the synergistic effects of the two drugs on packaged DNA inside cancer cells. What they found opens up possibilities for epigenetic targeting and suggests that allosteric modulation in nucleosomes may have biological relevance and potential for therapeutic interventions.
Detailed findings appeared March 30 in the journal Nature Communications, in an article entitled “Allosteric Cross-Talk in Chromatin Can Mediate Drug–Drug Synergy.” According to this article, drug–drug synergy between RAPTA-T and auranofin is mediated by allosteric cross-talk in chromatin.
“We found…RAPTA-T and auranofin…yield a synergistic activity in killing cancer cells, which coincides with a substantially greater number of chromatin adducts formed by one of the compounds when adducts from the other agent are also present,” wrote the article’s authors. “We show that this occurs through an allosteric mechanism within the nucleosome, whereby defined histone adducts of one drug promote reaction of the other drug at a distant, specific histone site.”
Essentially, the study found that combining the two drugs had an increased effect of killing of cancer cells, while individually, the drugs have considerably less impact on cell viability. When RAPTA-T is given, it forms adducts that disrupt the normal function of DNA and cause the cell to die. RAPTA-T also helps the other drug's ability to form histone adducts by binding on distant histone sites.
These results are particularly significant because they extend the scope of known allosteric interactions beyond protein-signaling systems, such as G protein-coupled receptors and different kinases, and drug-metabolizing enzymes, such as the P450 cytochromes. Until now, instances of allosteric drug activities on the histone protein-packaged form of cellular DNA, chromatin, have been unclear, and chromatin drug–drug interactions of allosteric origin have not been reported.
Despite popular depictions, the long strands of DNA in the cell spend most of their time tightly wound around specialized proteins called histones. Whenever a particular sequence, for example, a gene, is needed, that section of DNA is unwound and read by the appropriate biological machinery. A structural unit called the nucleosome is formed when eight histone protein cores serve as spool around which a segment of DNA is wound.
In the current study, the investigators speculated that the drug–drug interaction occurred at the nucleosome level: “[There] are likely to be multiple allosteric mechanisms within the nucleosome, which have the potential to modulate chromatin activity and can be influenced by even subtle chemical changes to the histone proteins.
“The allosteric mechanism at play could be dependent on the specific nucleosome binding event or alteration in question.” Of the possible kinds of alteration, the investigators lean toward the “domino” model, “a sequential set of local events propagates via a well-defined pathway from one allosteric active site to the other, spatially distant, site through a network of highly correlated neighbors.”
“The allosteric mechanism mediating drug–drug synergy we have characterized in this work suggests that there is untapped potential for therapeutic modulation of chromatin activity through exploitation of structural and dynamical features of the nucleosome,” the authors of the Nature Communications article concluded. “Although we have made the initial discovery here with metal-based drugs, which are especially favorable for structural visualization and cellular tracking, it is likely that synergies and allosteric actions in the nucleosome occur with other drug classes as well.”