The quest for new viral disease therapies and test kits has placed greater emphasis on RNA isolation and extraction. New technologies have been introduced that offer greater
simplicity, cost savings, and product availability over commercial kits, while achieving equivalent RNA quality and quantity. These technologies use single devices in addition to the laboratory’s leftover or preferred reagent kits.
One such technology, the nucleic acid extraction filter plate, can deliver high-quality isolation and purification of RNA and genomic DNA from mammalian cells at high throughput without sacrificing results. In cases where nucleic acid preparations require low to medium throughput, users frequently must employ kits containing expensive centrifugal devices that generate leftover reagent waste. Another new technology, the nucleic acid extraction spin column, can enable one device to perform these experiments with the laboratory’s own reagents.
High-throughput RNA purification with a nucleic acid extraction filter plate
The transcription of genes into RNA is an important step in the synthesis of functional gene products, which can be functional RNA species themselves or protein products formed after translation of messenger RNAs.
In many high-throughput gene expression analysis studies, cultured mammalian cells are exposed to different growth conditions. Reverse-transcription quantitative PCR (RT-qPCR) is used to measure the influence of the various conditions on the expression of genes of interest. Viral RNA isolation and extraction are essential prior to RT-qPCR in the detection of a virus in a sample.
This workflow is commonly performed in the development of vaccines and test kits for viral diseases. The studies are greatly aided by the availability of multiwell nucleic acid extraction filter plates with silica-based media that permit high-throughput total RNA isolation in a robust fashion.
Our study evaluated the performance of extraction filter plates versus commercially available RNA isolation kits. The filter plate contains a silica-based quartz glass fiber media for the isolation of total RNA from cultured mouse bEnd.3 endothelioma cells.
Robustness of the nucleic acid extraction filter plate as an RNA isolation medium was shown through the use of a protocol that relies on standard reagents readily obtained and prepared in-house by the researcher in an economically favorable manner. The latter protocolwas also implemented in an industrial process to isolate RNA from
mouse embryonic fibroblasts at a range of cell concentrations, and to
subject this RNA to one-step RT-qPCR assays for three genes.
Filter plate results
RNA isolation performance from cultured mouse bEnd.3 endothelioma cells was evaluated over a range of cell amounts employing two isolation protocols: the first with commercial reagents and the second with standard reagents prepared in-house.
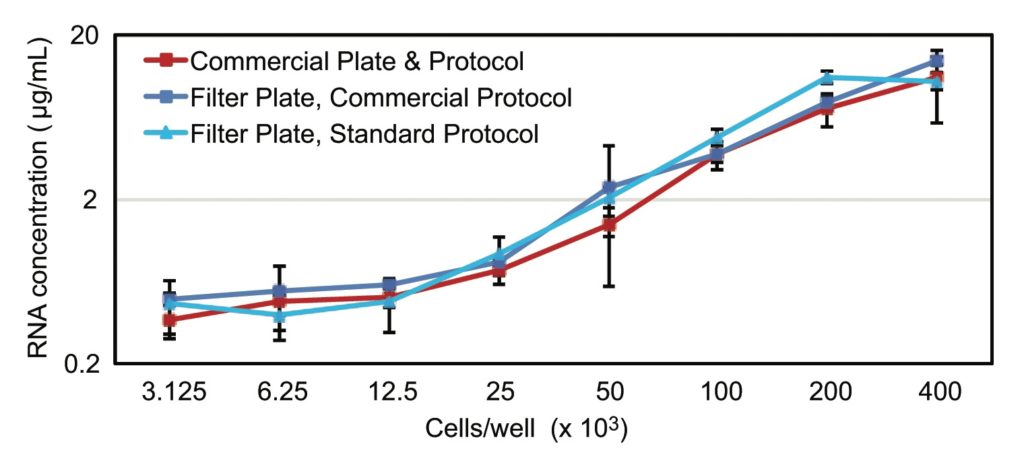
Results of the RNA concentration measurements using the Quant-iT RiboGreen RNA assay are shown in Figure 1. The results indicate that regardless of whether commercial reagents or standard reagents were used with the nucleic acid extraction filter plate, RNA yields were similar to those obtained with the commercial kit.
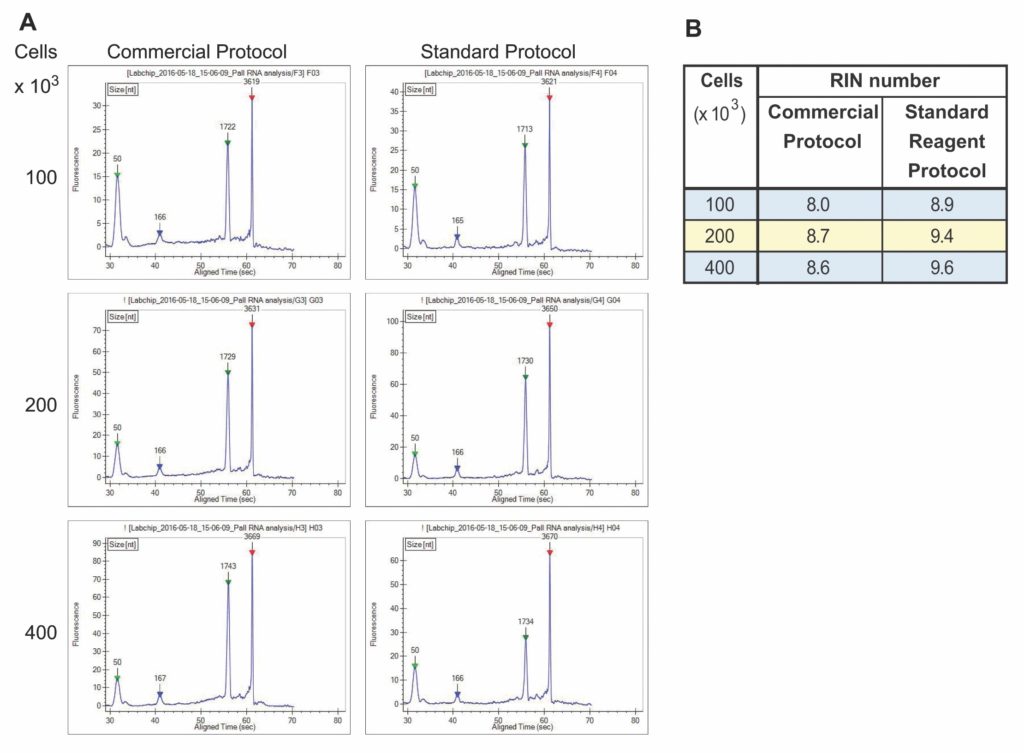
RNA samples isolated with the nucleic acid extraction filter plate were separated on a DNA 5K/RNA/CZE LabChip in the LabChip GX II Touch HT instrument to determine sample integrity. The instrument’s GX Touch software further assessed RNA integrity by performing a smear analysis of the electropherogram traces. An internal algorithm was used to calculate the RNA Integrity Number (RIN). The electropherograms showed two distinct RNA peaks corresponding to 18S and 28S rRNA with little to no evidence of a smear to indicate RNA degradation (Figure 2A). For samples derived from 100–400 × 103 cells, RIN numbers varied from 8.0 to 9.6 (Figure 2B), indicating that the RNA was of high quality.
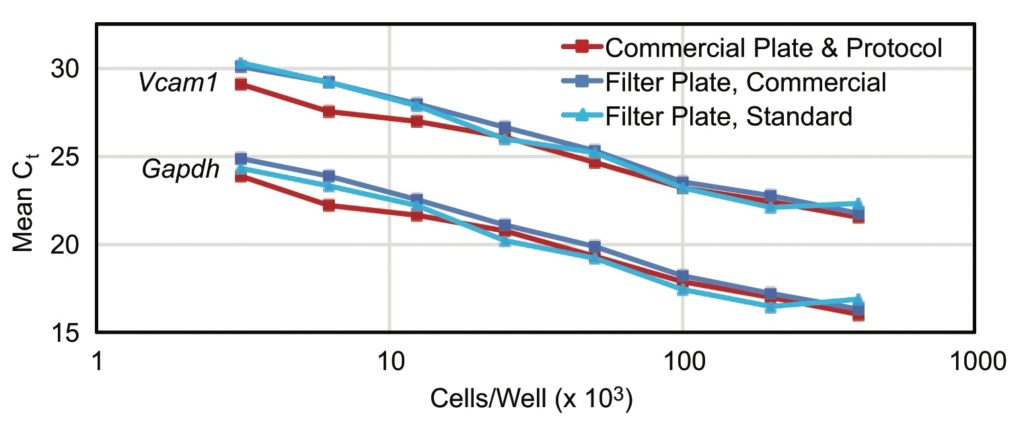
RNA integrity alone does not guarantee successful amplification in an RT-qPCR application as copurified inhibitors can impede reverse transcription and/or PCR efficiency. Mouse bEnd.3 cells are characterized by the expression of VCAM-1 (vascular cell adhesion molecule 1, which is encoded by the Vcam-1 gene). The average Ct values showed a
linear inverse relationship with increasing bEnd.3 cell numbers. In samples
isolated with either protocol or plate, both the abundant GADPH (housekeeping protein glyceraldehyde-3-phosphate dehydrogenase, which is encoded by Gapdh) and the less abundant VCAM-1 message were detected. The slopes of the curves through points of RNA samples isolated from increasing numbers of cells were equal, indicating that no
inhibitory components copurified.
Nucleic acids prepared with the extraction filter plate technology were of high quality and suitable for common downstream activities such as RT-qPCR, restriction digests, and sequencing (Figure 3).
Low- to medium-throughput RNA purification with a nucleic acid extraction spin column
The nucleic acid extraction spin column purifies RNA and genomic and plasmid DNA from bacteria, yeast, mammalian cultured cells, and plants. It is outfitted with an innovative duallayer, silica-based, quartz glass fiber matrix that enables its use with the buffers available in most currently available commercial kits.
Solid-phase extraction such as the spin-column-based method allows the nucleic acid to bind to the solid-phase matrix,1,2 limiting the problems associated with liquid-liquid
extraction.3 Solid-phase extraction facilitates the nucleic acid extraction process, making it fast, efficient, and reproducible compared with conventional methods.2,3 The extraction device recovers RNA fragments ranging in size from 50 bp to 10,000 bp.
Spin column results
Reagents from commercially available kits (CK1) were used, and RNA from Escherichia coli bacterial cultures, mammalian cultured cells (CHO), and plant leaves (basil) were extracted and purified with the spin column. As shown in Figure 4, the nucleic acid yields recovered with the extraction spin column were comparable to those obtained with the spin devices from commercially available kits.
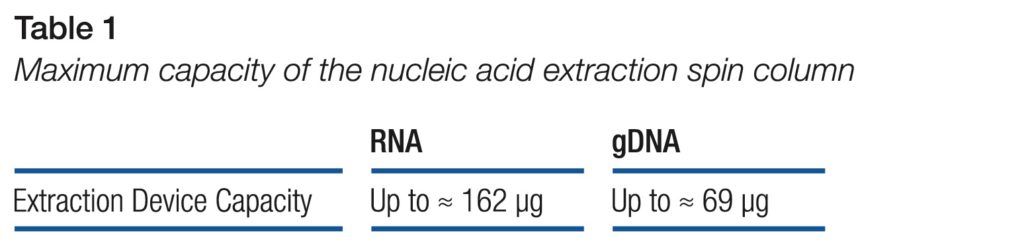
The data shows that the nucleic acid extraction spin column effectively extracts and purifies RNA and genomic DNA from common starting materials (Table 1).
The spin column has been thoroughly tested to assess performance with RNA extraction and purification from various starting materials. It proves that RNA (and genomic DNA) from bacteria, mammalian cells, and plants can be purified with this single device.
Moreover, the purified nucleic acids were processed in the most common molecular biology applications. The results demonstrated that nucleic acids purified with the extraction spin device meet the yield and purity requirements of the downstream applications.
Conclusions
The nucleic acid extraction filter plate and nucleic acid extraction spin column offer complementary options for high- and low-throughput nucleic acid purification. They are standalone, low-cost alternatives to purchasing full commercial kits. The technologies enable laboratories to perform RNA extraction and isolation with their own reagents and receive the RNA quantity and quality associated with commercial kits. These options are highly attractive at a time when viral research and development has placed a
premium on material availability.
Lori Euler ([email protected]) is the product manager for the molecular portfolio at Pall.
References
1. Ali N, Rampazzo RCP, Costa ADT, Krieger MA. Current Nucleic Acid Extraction Methods and Their Implications to Point-of-Care Diagnostics. Biomed. Res. Int. 2017; 2017: 9306564. DOI:10.1155/2017/9306564.
2. Zwir-Ferenc A, Biziuk M. Solid phase extraction technique: Trends, opportunities and applications. Pol. J. Env. Stud. 2016; 15(5): 677–690.
3. Tan SC, Yiap BC. DNA, RNA, and protein extraction:The past and the present. J. Biomed. Biotechnol. 2009; 2009: 574398. DOI: 10.1155/2009/574398.