Next-generation sequencing (NGS) is an imperfect science. Inherent biases and errors are serious considerations that affect the reliability of results and the bottom line. Current NGS offerings have improved only incrementally over the years, leaving advancements in sample preparation as the sole way to elevate efficiency.
Target enrichment is a way to achieve more focused sequencing, with increased precision and savings in sequencing costs. However, the use of low-quality probes and suboptimal protocols result in the need for higher read depths to cover target sequences, which can considerably increase costs. In addition, standardized metrics to evaluate sequencing and target enrichment performance have yet to be fully realized, leaving users of the technology unsure how to improve results.
Twist Bioscience has focused on designing and manufacturing high-quality target enrichment probes, and on adding unique twists on established NGS protocols to deliver an NGS workflow with uniform and deep coverage. With a focus on uniform target capture, and a unique dsDNA probe design, confidence in sequencing reads is maximized, while minimizing sequencing costs. The results discussed in this article demonstrate that the methods Twist uses and recommends for NGS target enrichment are more efficient than other established protocols.
Coverage uniformity has a greater impact on efficient target capture
Although targeted NGS can reduce sequencing costs and make experiments more feasible, target enrichment techniques can introduce biases that compromise efficiency and lead to “wasted sequencing.” Inefficient target capture, due to suboptimal probes, results in some areas with low read depth which require increased sequencing to ensure high-
confidence data. This leads to oversequencing of otherwise adequately covered regions and higher sequencing costs. Efficient target capture, using quality probes, is the key to reducing wasted sequencing.
The on-target rate and coverage uniformity are two metrics of probe panels that affect target capture efficiency and should be considered when choosing probes for target enrichment. The on-target rate refers to the percentage of probes that exclusively hybridize with, and capture, targeted regions. Coverage uniformity refers to the uniformity of target capture among the probe panel and can be expressed as the fold-80 base penalty (fold-80), calculated by the widely used Picard pipeline. Fold-80 is defined as the fold of additional sequencing required to ensure that 80% of the target bases achieve the mean coverage. The lower the on-target rate, or the higher the fold-80, the higher the capture inefficiency and wasted sequencing.
While uniformity and on-target rates are important metrics to achieve efficient sequencing, small improvements in uniformity have a much larger impact on increasing efficiency. Figure 1 shows the impact of fold-80 and the on-target rate on the proportion of target bases covered at 10× coverage or higher. Each colored curve represents a different fold-80, ranging from 1.1 to 2.0. The vertical thickness of each line represents the change in the on-target rate from 80 to 100%. For example, at 30× mean coverage—when fold-80 is fixed at 1.7 (blue curve)—an increase in the on-target rate from 80 to 100% results in an increase in the proportion of target bases covered by about 3%. In contrast, when the on-target rate is fixed at 100% (the top of curves), an increase in fold-80 from 1.7 to 1.4 (blue to green curve) results in an increase in the proportion of target bases covered by about 6%.
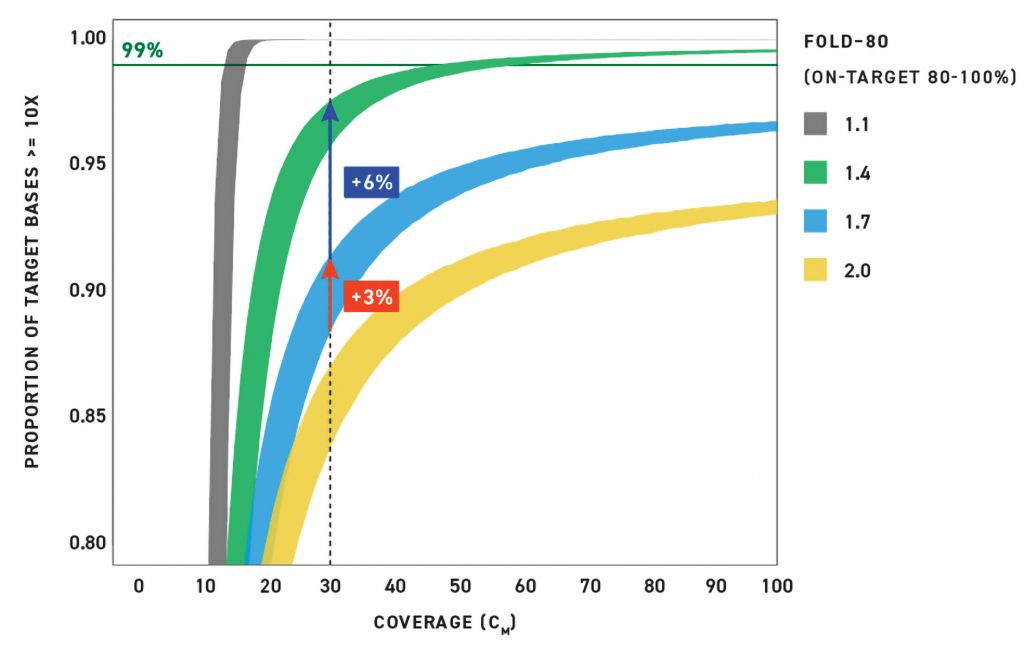
This model demonstrates that coverage uniformity has a much higher impact on the efficacy of targeted sequencing. Given this fact, Twist recommends that, in addition to the on-target rate, coverage uniformity should be carefully considered when choosing probe panels for target enrichment. Twist uses design and manufacturing protocols to maximize coverage uniformity and the on-target rate of its probes to maximize capture efficiency and minimize wasted sequencing.
Double-stranded DNA probes elevate sequencing accuracy and uniformity
Traditionally, probes in double-stranded form have been thought to bind with each other and compete against target-probe hybridization. Twist, however, has developed a proprietary method that can enable the use of dsDNA probes without compromising target capture. When both strands of the target are specifically captured, an increased number of unique molecules can be sequenced, enabling the detection of strand-specific mutations, such as deamination events of partially degraded samples, which result in higher library diversity and fewer PCR duplicates for high-confidence NGS. dsDNA probes increase NGS accuracy and sensitivity by differentiating between rare mutations and nucleotide changes due to processing or degradation—an advancement critical to cancer sequencing.
Besides dsDNA probes, efficient target capture requires good design and high-quality manufacturing of probe oligos. Through extensive experimentation, machine learning, and design-test-learn cycles, Twist has developed advanced algorithms to design its NGS probes. These algorithms are tailored to consider not only the target organism and panel being designed, but also Twist’s manufacturing process and target enrichment workflow, thereby yielding improved uniformity and lower off-target rates.
If seed oligos are amplified with standard PCR, biases will be introduced based on their GC content. This results in an unbalanced capture pool, which leads to nonuniform target capture. Twist takes a different approach to oligo amplification, which maintains the uniformity of the seeds within the final oligo pools. Each oligo is synthesized individually using Twist’s high-throughput silicon platform to achieve amplification, without the bias of PCR. The resulting uniform capture probe pools allow for more uniform sequencing, and with less waste. Using this manufacturing technology—paired with Twist’s probe design algorithms—probe pools are on-target with highly uniform capture.
To test the sequencing efficiency of Twist’s dsDNA probes in real-world settings, two independent studies, one by Johns Hopkins University1 and one by CeGaT,2 compared the performance of the Twist Human Core Exome Kit with other commercially available exome enrichment protocols. Each study used independent metrics to assess performance, and data from both studies showed that the Twist Human Core Exome Kit was superior to its competitors in its ability to generate high-quality data at lower sequencing cost.
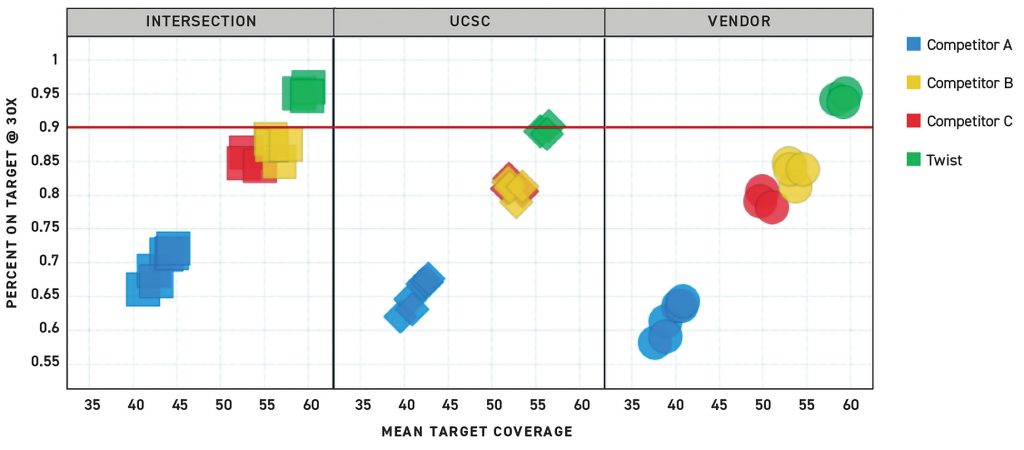
Figure 2 shows representative data from the John Hopkins University study illustrating that only samples prepared with the Twist Human Core Exome Kit achieved >90% on-target rate at 30× coverage. CeGaT also assessed the uniformity, specificity, and complexity offered by a similar set of exome enrichment protocols and found that Twist’s Human Core Exome Kit exhibited the highest efficiency with the lowest input requirements. Using the Twist kit also resulted in more efficient read usage and higher library complexity, as reflected in the extremely low duplicate rate of <5% at 12 Gb sequencing output. In contrast, the other enrichment methods resulted in more than three times that rate.
References
1. Marosy B, Gearhart J, Craig B, Doheny KF. Comparison of Whole Exome Capture Products—Coverage & Quality vs Cost. Poster paper presented at the ASHG 2018 Annual Meeting (PgmNr 1795).
2. CeGaT. On the quest for a more precise exome.






