The past few years have seen an explosion in interest in immuno-oncology approaches that leverage monoclonal antibodies (mAbs) as therapeutics for a range of cancers. These therapies take advantage of the ability of antibodies to trigger antibody-dependent cellular cytotoxicity (ADCC) within the innate immune response, in which the patient’s immune cells (predominantly natural killer T cells) respond to the binding of the mAb to antigens on the surfaces of cancer cells by actively lysing the cancer cells.1,2
Recent research has shown that the glycosylation state of therapeutic antibodies––particularly fucosylation––can modulate the activity of the antibodies and thus the robustness of the ADCC response. The Fc region of mAbs functions as a critical bridge between the adaptive and innate immune response, notably including the activation of ADCC via natural killer Tcells.
More precisely, the removal of fucosylation present on the Asn297 glycan opens the Fc conformation of immunoglobulin G1 (IgG1), leading to stronger affinity to the FcγRIIIa present on natural killer T cells. When applied to anticancer mAbs, research has demonstrated that removal of fucose moieties from the IgG1 Fc region provides an effective means of increasing ADCC activity, and hence more potent antibodies, driving the active killing of cancer cells.3–6
Following on these findings, there are currently three afucosylated antibodies on the market, with over 20 currently being investigated in clinical trials.7 While glycoengineering to reduce the fucosylation of core Fc residues appears to be a reliable strategy to increase the robustness of mAb therapies for oncology applications, expression systems that incorporate glycoengineering tools to evaluate the importance of N-glycosylation profiles in the target product profile often lack consistency and scalability. Here, we present a more comprehensive toolbox that allows the screening of different glycoforms at the discovery stage using a system that can support development and manufacturing in a rapid and cost-effective manner.
Current glycoengineering limitations
Although glycoengineering can be leveraged later in the workflow as a means of optimizing a selected mAb, the entire development process could be streamlined and made more efficient by incorporating a comprehensive glycoengineering strategy earlier in the screening process, especially if it can be implemented in a reproducible and scalable manner. While Chinese hamster ovary (CHO) and Pichia pastoris expression systems can be modified to modulate glycosylation patterns, this is typically performed by altering nutrients in the growth media and does not present clear pathways to scale-up.
Plant-based solution
Some alternative protein expression systems, such as iBio’s FastPharming® platform utilizing Nicotiana benthamiana plants, enable glycosylation engineering by design at pilot scale that can be rapidly scaled for clinical and commercial production.
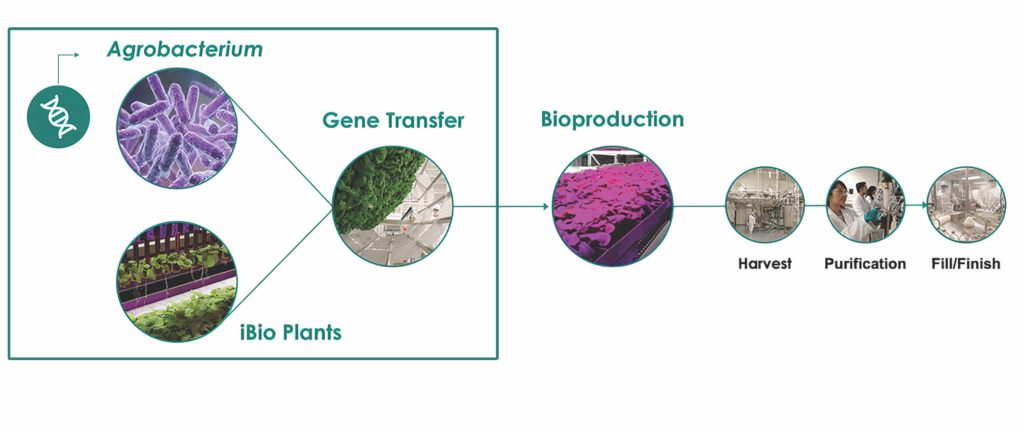
Under the FastPharming system, plants are used for the transient expression of recombinant proteins at scale. The gene of interest is transfected into plant leaves via agroinfiltration, which presents a linear and scalable process that bypasses the need for cell line development and extensive tank bioreactor scale-up studies (Figure 1). The target protein is expressed in the leaves as the plants grow. The leaves are then harvested and the protein isolated, purified, and formulated using traditional pharmaceutical methods.
Scale-up is achieved by simply growing more plants for transfection and maintaining highly reproducible process parameters during process development, whether at clinical or commercial scale. Importantly, control of post-translational modifications is greater
in this plant-based expression technology because the size of the bioreactor (a single plant) does not change as the upstream process scales up, leading to increased batch-to-batch consistency and product quality.
In addition, protein N-glycosylation in plants tends to be more homogeneous because plants do not perform sialylation, α-(1,3) galactosylation, or β-(1,4) galactosylation in wild-type backgrounds. This particular feature opened the opportunity to design transgenic hosts that offer tighter N-glycosylation control and that can be used from discovery to manufacturing for the production of glycoengineered proteins.
Customized glycoengineering
Using FastPharming, iBio designed glycoengineering tools to offer a range of custom N-glycosylation options adapted to enhance utility in biotherapeutic applications (Figure 2). Wild-type plants naturally produce complex N-glycans deprived of galactosylation and sialylation but containing α-(1,3) fucose and β-(1,2) xylose. By inactivating the plant-specific glycosyltransferase, a ΔXT/FT plant produces a very homogeneous G0 and
afucosylated glycoform (Figure 3). By further engineering the plant, the knock-in of the human α-(1,4) galactosyltransferase allowed the generation of G1 and G2 afucosylated proteins.
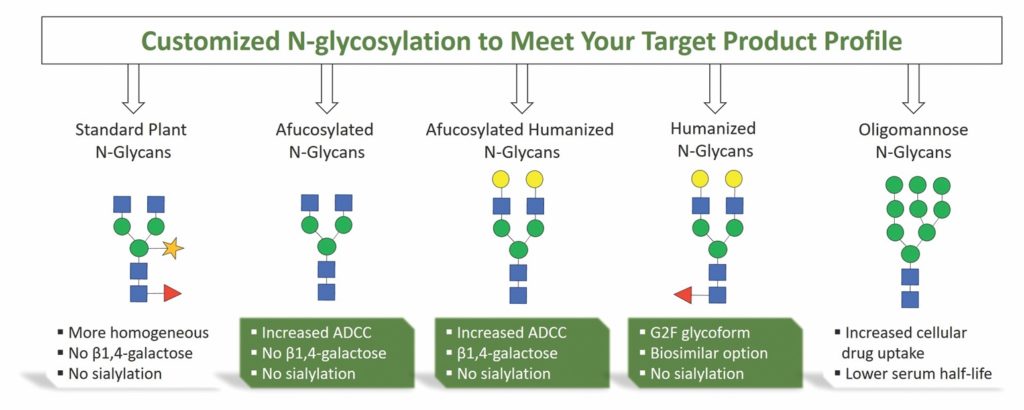
By expressing target proteins in those different plant backgrounds, one leverages the power of FastPharming with customized glycoengineering, leading to early decision points in the discovery program and rapid escalation to preclinical and clinical development. The ability to develop a range of products with targeted glycosylation patterns allows for efficient screening of different glycoforms with respect to ADCC response.
The potential of FastPharming for glycoengineering was demonstrated using rituximab, focusing on the production of afucosylated forms. First, G0 rituximab was generated using a transgenic plant engineered to remove the core fucose. With a second transgenic plant expressing the human β-(1,4) galactosyltransferase, G1 and G2 afucosylated rituximab glycoforms were produced. All proteins were produced in parallel using a readily scalable process. In addition to eliminating antibody fucosylation, this approach led to a significant reduction of the glycosylation profile complexity overall, facilitating further development and manufacturing efforts.
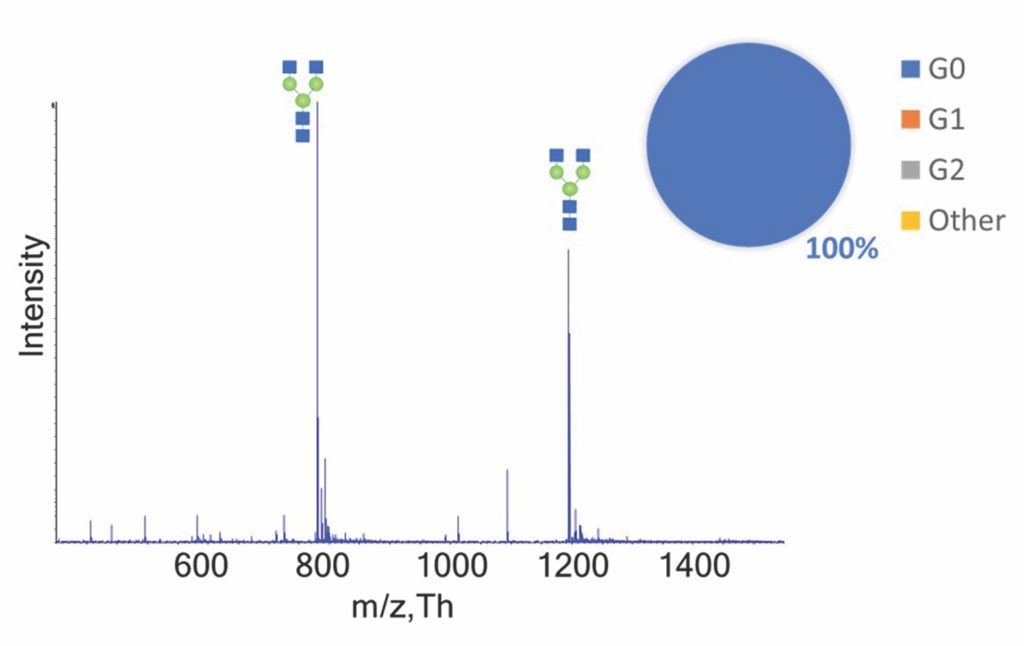
As can be seen in Figure 3, rituximab derived from the ∆XT/FT plants comprised the homogeneous G0 glycoform. However, the ∆XT/FT + GalT plants generated G1/G2 glycoforms comparable to those obtained from cell culture using FUT8−/− CHO knockout cell lines (data not shown).
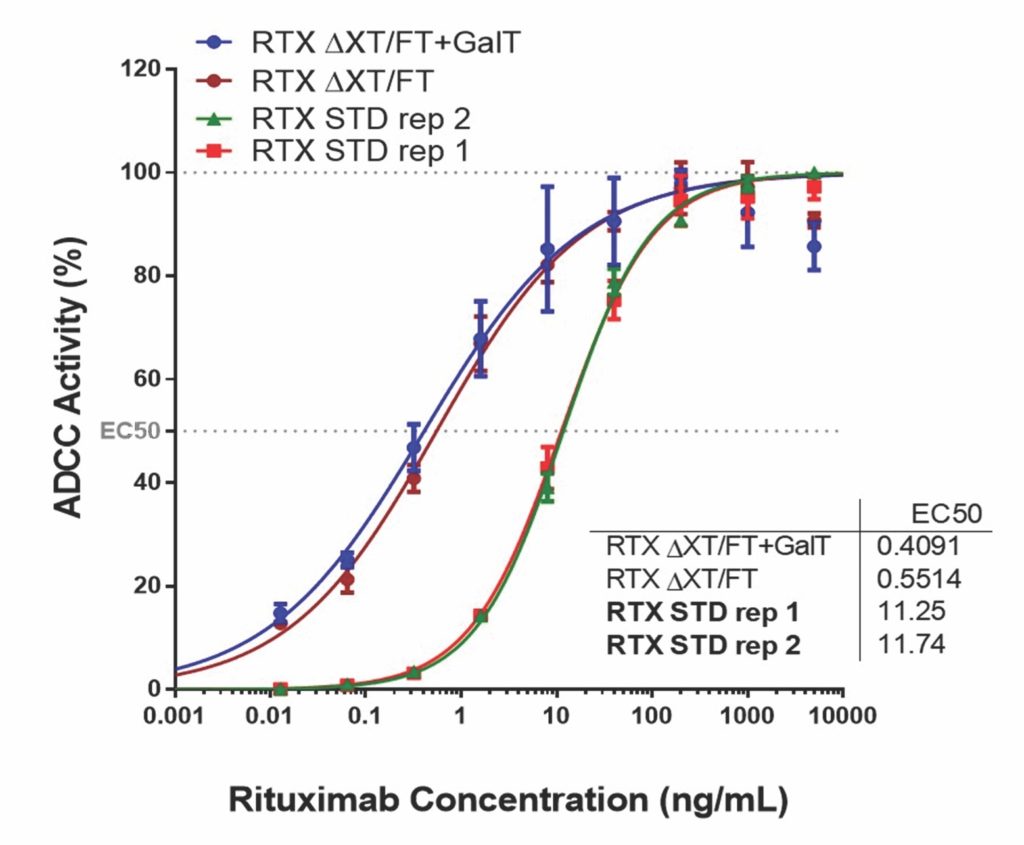
All of the glycoengineered rituximab proteins exhibited enhanced ADCC activity in vitro (Figure 4) without affecting the protein’s ability to bind the CD20 receptor and other key properties (data not shown).
Comprehensive glycoengineering services
Currently, iBio is enhancing its Glycaneering™ Development Service and expects the service improvements to be available in the second half of 2021. Provided with a specific sequence, iBio will use its toolkit of different plants and methods of expression to uniformly produce specific glycoforms for activity screening within five weeks (lab scale). Scale-up to gram quantities for comparability and confirmation studies can follow rapidly.
Furthermore, combining Glycaneering with multiattribute analytical methods for rapid protein/antibody characterization will provide a comprehensive data package on the different glycoforms that allows data-driven decision making, which is critical for selection of the optimum protein or antibody to advance the drug development process.
Sylvain Marcel, PhD, is vice president of protein expression sciences at iBio.
References
1. Cartron G, Dacheux L, Salles G, et al. Therapeutic activity of humanized anti-CD20 monoclonal antibody and polymorphism in IgG Fc receptor FcgammaRIIIa gene. Blood 2002; 99: 754–758. DOI: 10.1182/blood.V99.3.754. PMID: 11806974.
2. Weng WK, Levy R. Two immunoglobulin G fragment C receptor polymorphisms independently predict response to rituximab in patients with follicular lymphoma. J. Clin. Oncol. 2003; 21: 3940–3947. DOI: 10.1200/JCO.2003.05.013. PMID: 12975461.
3. Shields RL, Namenuk AK, Hong K, et al. High resolution mapping of the binding site on human IgG1 for Fc gamma RI, Fc gamma RII, Fc gamma RIII, and FcRn and design of IgG1 variants with improved binding to the Fc gamma R. J. Biol. Chem. 2001; 276: 6591–6604. DOI: 10.1074/jbc.M009483200. PMID: 11096108.
4. Iida S, Misaka H, Inoue M, et al. Nonfucosylated therapeutic IgG1 antibody can evade the inhibitory effect of serum immunoglobulin G on antibody-dependent cellular cytotoxicity through its high binding to FcgammaRIIIa. Clin. Cancer Res. 2006; 12: 2879–2887. DOI: 10.1158/1078-0432.CCR-05-2619. PMID: 16675584.
5. Shields RL, Lai J, Keck R, et al. Lack of fucose on human IgG1 N-linked oligosaccharide improves binding to human Fcgamma RIII and antibody-dependent cellular toxicity. J. Biol. Chem. 2002; 277: 26733–26740. DOI: 10.1074/jbc.M202069200. PMID: 11986321.
6. Shinkawa T, Nakamura K, Yamane N, et al. The absence of fucose but not the presence of galactose or bisecting N-acetylglucosamine of human IgG1 complex-type oligosaccharides shows the critical role of enhancing antibody-dependent cellular cytotoxicity. J. Biol. Chem. 2003; 278: 3466–73. DOI: 10.1074/jbc.M210665200. PMID: 12427744.
7. Pereira NA, Chan KF, Lin PC, Song Z. The “less-is-more” in therapeutic antibodies: Afucosylated anti-cancer antibodies with enhanced antibody-dependent cellular cytotoxicity. MAbs 2018; 10(5): 693–711. DOI: 10.1080/19420862.2018.1466767. PMCID: PMC6150623. PMID: 29733746.

