Proteolysis-targeting chimeras (PROTACs) are bifunctional molecules that hijack ubiquitin E3 ligases and induce degradation of intracellular proteins through a tightly regulated proteasomal mechanism. Although several successful PROTACs have been developed against key intracellular target classes including bromodomains, kinases, and nuclear hormone receptors, these bivalent molecules often suffer from poor cell permeability due to high molecular weight.
To enable a high-throughput, quantitative readout for PROTAC cell permeability and E3 ligase occupancy in living cells, we have developed a panel of NanoBRET™ target engagement (TE) assays for key E3 ligases or adapter proteins including CRBN, VHL, XIAP, cIAP, and MDM2. The NanoBRET TE intracellular assays are the first biophysical method to enable the quantitative determination of compound occupancy, potency, and residence time for specific target proteins inside living cells using bioluminescent resonance energy transfer (BRET). This method has previously been applied to several other protein classes including kinases, bromodomains, and HDACs (Figure 1).
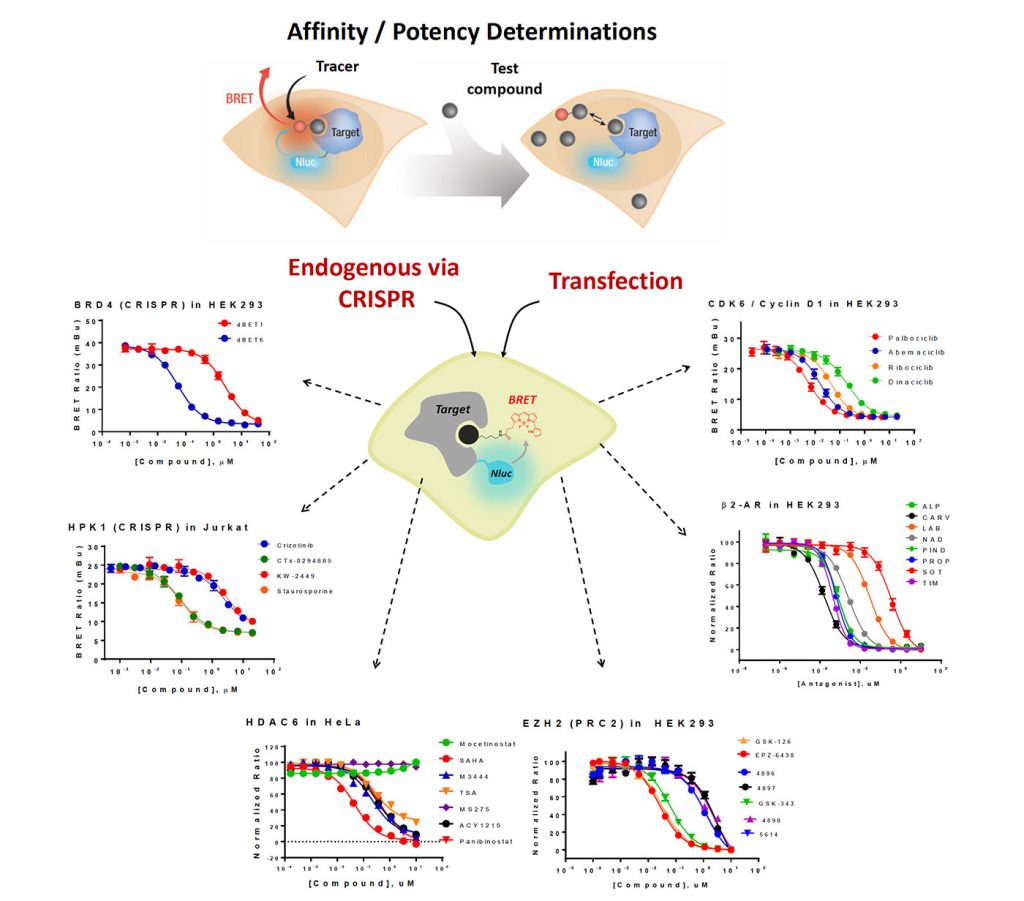
Here, we demonstrate that the NanoBRET platform allowed assessment of PROTAC cell permeability and E3 ligase occupancy. Using NanoBRET TE assays for proteins targeted by the E3 ligases, we obtained intracellular PROTAC binding kinetics. We further extend the analysis of PROTAC permeability as a dynamic process in real time, using BRD-targeting MZ1 and dBET1 as a model system. Together, these approaches allow a mechanistic interrogation of intracellular PROTAC permeability, E3 ligase occupancy, target occupancy, and target
residence time.
Quantitate molecular glue and PROTAC occupancy and affinity at CRBN
The TE assays can be run in live or permeabilized cells (Figure 2A), enabling assessment of compound intracellular affinity and permeability. Relative shifting in IC50 between live and permeabilized formats can reveal the impacts of chemical structure on PROTAC permeability.
The live-cell assay was used to quantify the apparent intracellular affinity of a series of small-molecule CRBN molecular glues (Figure 2B). With permeabilized cells, there was little change in IC50 observed compared to when the assay was run in live cells, indicating these compounds were readily cell permeable (Figure 2B).
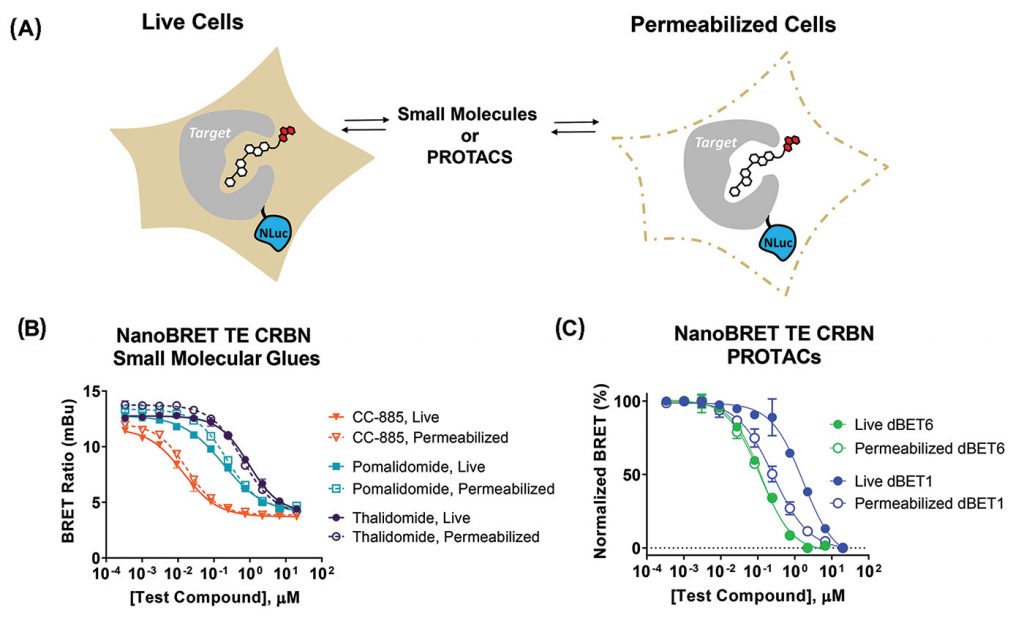
In contrast to small molecular weight glues, bifunctional PROTACs may have limited cellular permeability. To understand PROTAC permeability, TE assays can be run in live cells or permeabilized cells (Figure 2C). Comparing results from a TE CRBN assay in live cells to permeabilized cells revealed that (1) dBET6 is more permeable than dBET1 and (2) dBET6 is slightly more potent than dBET1.
Measure MZ1 permeability using VHL and BRD4 target engagement assays
BRD4 or VHL NanoLuc fusions were individually expressed in HEK293 cells. Intracellular engagement was quantified using TE BRD Tracer 02 or VHL tracer, following a 3-hr equilibration (Figures 3A & 3B). The offset in engagement potency of PROTAC MZ1 compared to precursors VH298 or JQ1 may suggest relatively weaker permeability of the PROTAC MZ1.
Engagement potency can also be measured in real time against BRD4 (Figure 3C). Plotting compound IC50 vs. time reveals slow equilibration of PROTACs MZ1 and dBET1 to BRD4 compared to JQ1 precursor, supporting comparatively higher permeability in live cells for JQ1.
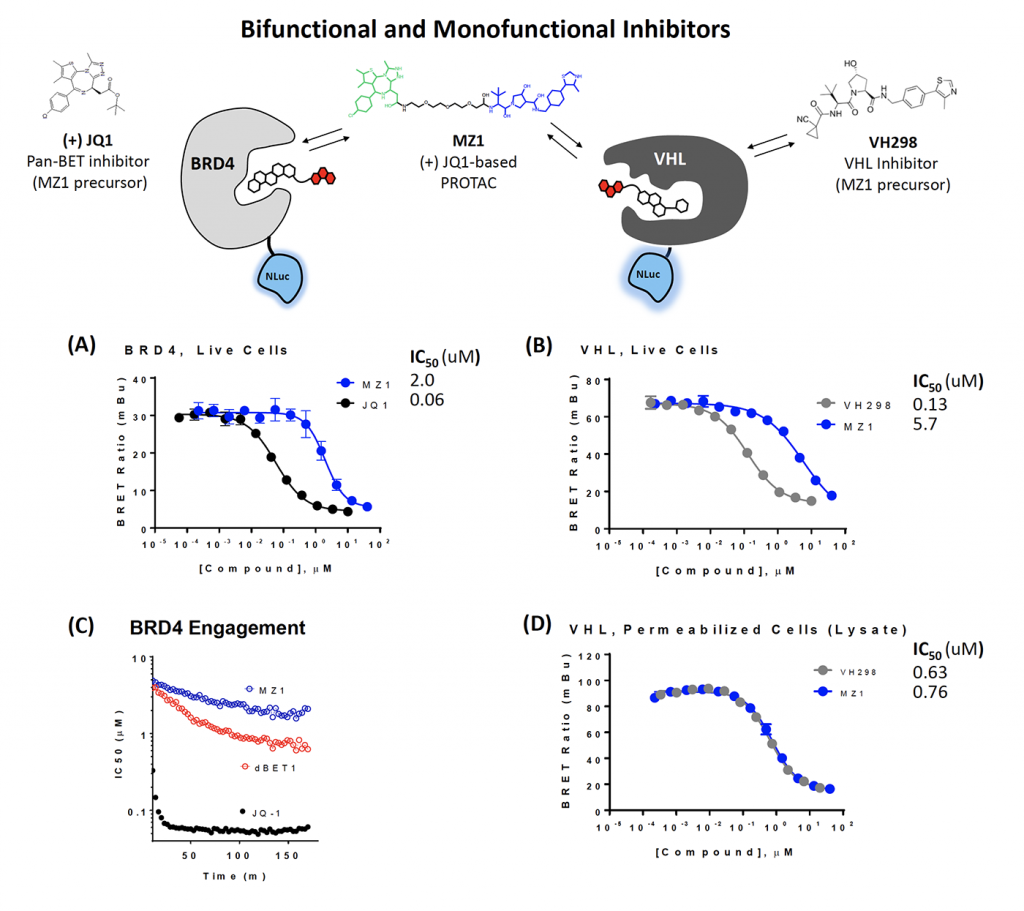
NanoBRET can also be performed in permeabilized cells to examine the impact of PROTAC permeability. In live cells, the MZ1 PROTAC shows right-shifted IC50 compared to precursor VH298 (Figure 3B). When cells are permeabilized, the potency values are identical (Figure 3D). This supports the relatively weak permeability of the PROTAC compared to the VH298 parental inhibitor.
Quantitate small-molecule inhibitor affinity for XIAP, clAP1, and MDM2 in live cells
Inhibitor of apoptosis (IAP) family proteins are E3 ligases and are frequently overexpressed in cancer. Development of inhibitors that can induce degradation of IAP family members may be of therapeutic interest. TE assays have been developed for two of the IAP family members, XIAP and cIAP1. Intracellular affinity for the IAP inhibitors BV-6 and LCL-161 were determined for both XIAP and cIAP1 in HEK293 cells (Figures 4A & 4B).
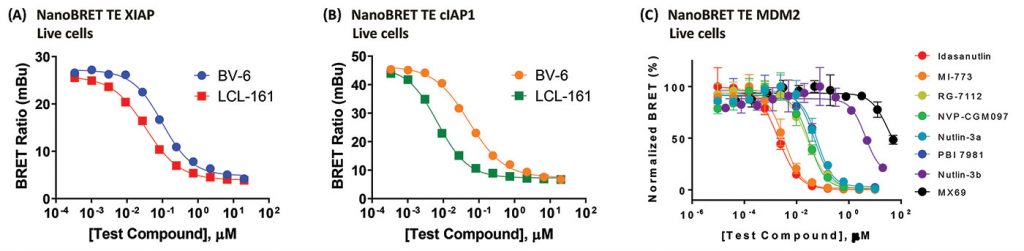
The E3 ligase MDM2 has many roles including the ubiquitination of the tumor suppressor p53, marking it for degradation by the proteasome. MDM2 is also overexpressed in many cancers. As a result, development of MDM2 inhibitors may be of clinical interest. Nutlins are small-molecule MDM2 inhibitors. Intracellular affinities of a series of nutlin derivatives for MDM2 were determined in HEK293 cells using TE following a 2-hr equilibration (Figure 4C).
Conclusions
TE assays broadly enable the quantitative determination of compound affinity/potency for specific targets inside cells. Cell-permeable NanoBRET tracers have been developed that allow TE assays for >200 full-length kinases and five E3 ligases. Furthermore, TE has been successfully used to interrogate live-cell compound engagement for hundreds of intracellular targets, spanning a variety of protein classes in the human proteome.
Cellular affinity, permeability, and occupancy of small-molecule inhibitors, molecular glues, and PROTACs for E3 ubiquitin ligases can be assessed via NanoBRET TE assays, including the E3 ligases or adapter proteins including CRBN, VHL, XIAP, cIAP1, and MDM2. TE assays can be run in live and lytic mode that aids in understanding compound permeability. Additionally, TE live-cell assays can be run in real time to allow examination of kinetics of intracellular binding.
The NanoBRET TE method should facilitate the development of PROTAC and E3 ligase inhibitors with optimal cell permeability and target occupancy.
James Vasta, PhD, is a senior research scientist and Matthew Robers ([email protected]) is a senior scientist and group leader at Promega.