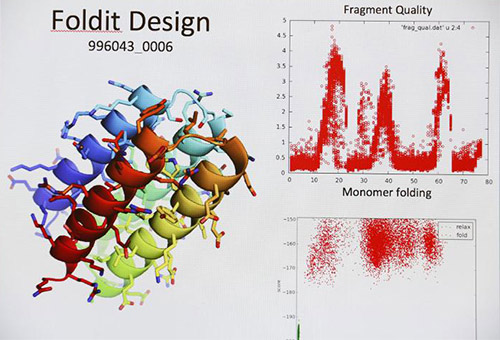
![Volunteer citizen scientists around the world participate in research on protein structure prediction through the computer program Foldit. [Institute for Protein Design/University of Washington]](https://genengnews.com/wp-content/uploads/2018/08/Dec17_2015_UnivWash_FolditDesign9119631212-1.jpg)
Volunteer citizen scientists around the world participate in research on protein structure prediction through the computer program Foldit. [Institute for Protein Design/University of Washington]
Although nature uses the modular approach to construct a dizzying array of proteins, it explores a fairly small region of protein sequence and structure space. Perhaps computers in the employ of protein scientists can be more thorough. Intending to explore strange new protein worlds—and to boldly go where nature itself has not gone before—protein scientists are using computational approaches to fully exploit the possibilities of modular assembly in protein space.
Scientists based at the University of Washington Institute for Protein Design and the Fred Hutchinson Cancer Research Institute in Seattle described their most recent voyages in a pair of papers published December 16 in Nature. The papers are “Exploring the repeat protein universe through computational design” and “Rational design of alpha-helical tandem repeat proteins with closed architecture.” These papers suggest that the possibilities for producing useful protein geometries far exceed what nature has achieved.
“[We used] computational protein design to investigate the space of folded structures that can be generated by tandem repeating a simple helix–loop–helix–loop structural motif,” wrote the authors of the “Exploring” article. “Eighty-three designs with sequences unrelated to known repeat proteins were experimentally characterized. Of these, 53 are monomeric and stable at 95 °C, and 43 have solution X-ray scattering spectra consistent with the design models.”
“[We used] computational protein design to investigate the space of folded structures that can be generated by tandem repeating a simple helix–loop–helix–loop structural motif,” wrote the authors of the “Rational design” article. “Eighty-three designs with sequences unrelated to known repeat proteins were experimentally characterized. Of these, 53 are monomeric and stable at 95 °C, and 43 have solution X-ray scattering spectra consistent with the design models.”
The authors of both articles concluded that their results showed that existing repeat proteins occupy “only a small fraction of the possible repeat protein sequence and structure space.” In addition, they expressed optimism about the possibilities of designing novel repeat proteins with precisely specified geometries. Their computer-guided approaches, they noted, could open a wide array of new possibilities for biomolecular engineering.
The researchers developed powerful algorithms to make unprecedented, accurate, blind predictions about the structure of large proteins of more than 200 amino acids in length. This work has made it easier to predict the structures for hundreds of thousands of recently discovered proteins in the ocean, soil, and gut microbiome.
Equally difficult is designing amino acid sequences that will fold into brand new protein structures. Researchers have now shown the possibility of doing this with precision for protein folds inspired by naturally occurring proteins. More importantly, researchers can now devise amino acid sequences to fashion novel, previously unknown folds, far surpassing what is predicted to occur in the natural world.
The new proteins are designed with help from volunteers around the globe participating in the Rosetta@home distributed computing project. The custom-designed amino acid sequences are encoded in synthetic genes, the proteins are produced in the laboratory, and their structures are revealed through X-ray crystallography. The computer models in almost all cases match the experimentally determined crystal structures with near atomic level accuracy.
“It has been a watershed year for protein structure predictions and design,” said UW Medicine researcher David A. Baker, UW professor of biochemistry, Howard Hughes Medical Institute investigator, and head of the UW Institute for protein design.
Researchers have also reported new protein designs, all with near atomic level accuracy, for such shapes as barrels, sheets, rings, and screws. This adds to previous achievements in designing protein cubes and spheres, and suggests the possibility of making a totally new class of protein materials.
By furthering advances such as these, researchers hope to build proteins for critical tasks in medical, environmental and industrial arenas. Examples of their goals are nanoscale tools that: boost the immune response against HIV and other recalcitrant viruses, block the flu virus so that it cannot infect cells, target drugs to cancer cells while reducing side effects, stop allergens from causing symptoms, neutralize deposits, called amyloids, thought to damage vital tissues in Alzheimer's disease, mop up medications in the body as an antidote, and fulfill other diagnostic and therapeutic needs. Scientists are also interested in new proteins for biofuels and clean energy.