If only the cell’s recycling components were as conspicuous as professional wrestlers. Then we might stand a better chance of understanding how they accomplish an essential task: the detection and degradation of misfolded proteins. Somehow, these warped, bent, or otherwise misshapen molecules are consigned to proteasomes, the cell’s ultimate recycling centers. But the steps between a protein’s acquisition of an unfortunate shape—whether by genetic mutation, synthesis gone awry, or simple wear and tear—and its proteolytic degradation remain unclear, even though they are known to stave off devastating neurodegenerative diseases such as Alzheimer’s and Parkinson’s.
It’s almost as though a championship match has been rumbling along, round by round, with much of the action happening out of sight. But recently researchers have managed to get ringside seats.
Xiaolu Yang, Ph.D., professor of cancer biology, Perelman School of Medicine at the University of Pennsylvania, together with his colleagues, identified a protein recycling pathway in mammalian cells that removes misfolded proteins. Their findings appeared online May 29 in Molecular Cell, in an article entitled “A Cellular System that Degrades Misfolded Proteins and Protects against Neurodegeneration.” As this title suggests, the article also describes how the researchers demonstrated this pathway’s role in protecting against neurodegenerative diseases.
“It is unclear how misshapen proteins are recognized and shuttled to the proteasome to be degraded,” noted Dr. Yang. Nonetheless, he expressed optimism that his team’s work, which uncovered key molecular players in protein recycling and their mechanism, would help resolve a long-standing question in protein quality control. “This study moves the field forward because we showed that the system is common for many types of misfolded proteins,” he added.
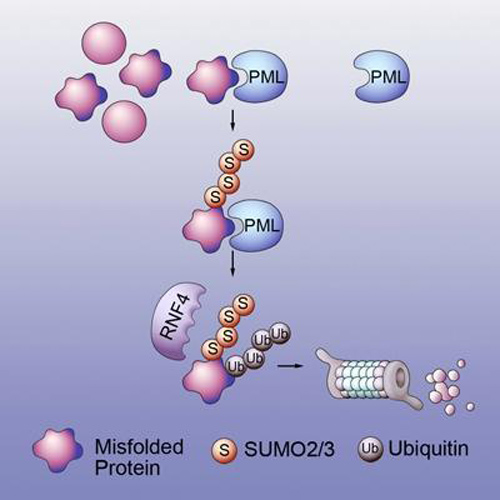
The mechanism of action uncovered by Dr. Yang and his colleagues is a relay system with two proteins. The first protein, PML/TRIM19, recognizes features of misfolded proteins such as exposed water-phobic inner cores, which stick out and enhance the forming of toxic protein clumps. PML selectively interacts with misfolded proteins “through distinct substrate recognition sites and conjugates these proteins with the small ubiquitin-like modifiers (SUMOs) through its SUMO ligase activity.”
The SUMO-modified misfolded proteins are then recognized and ubiquinated by the second protein, RNF4. The ubiquitin chain is a signal that can be recognized by the barrel-shaped proteasome, leading to the degradation of misfolded proteins.
The researchers then went on to demonstrate the physiological importance of the elimination system using a mouse model of spinocerebellar ataxia 1 (SCA1), a fatal neurological disorder that causes problems with movement and balance. Mutations in the ATXN1 gene cause SCA1, which involves repeated segments of the DNA building blocks cytosine (C), adenine (A), and guanine (G) that appear multiple times in a row in the gene, which encodes a tract of contiguous glutamine amino acids.
Normally, the CAG segment is repeated 4 to 39 times within the gene. In SCA1, the CAG segment is repeated 40 to more than 80 times, which leads to an abnormally long ataxin-1 protein that folds into the wrong three-dimensional shape and forms clumps within the nucleus.
The team showed that a PML deficiency exacerbates both the behavioral and neuropathological defects caused by the expanded CAG repeat in the SCA1 mouse, indicating that PML normally operates to protect against neurodegeneration.
“The knowledge gained from this project now provides for the possibility of new therapeutic targets for neurodegenerative diseases. Perhaps new drugs could enhance the elimination system by increasing the action of PML or RNF4, or could inhibit the inhibitors of the SUMO and ubiquitin tagging process,” concluded Dr. Yang.