An international research team headed by scientists at the University of Washington has successfully applied reinforcement learning, a strategy proven adept at board games like Chess and Go, to develop a powerful new protein design software. In one experiment, proteins made using the new approach were found to be more effective at generating useful antibodies in mice.
The work represents what they suggest is a milestone in tapping artificial intelligence to conduct protein science research. The potential applications could be wide ranging, from developing more potent vaccines and more effective cancer treatments, to creating new biodegradable textiles.
“Our results show that reinforcement learning can do more than master board games,” said David Baker, PhD, professor of biochemistry at the UW School of Medicine in Seattle and a recipient of the 2021 Breakthrough Prize in Life Sciences. “When trained to solve long-standing puzzles in protein science, the software excelled at creating useful molecules. If this method is applied to the right research problems. It could accelerate progress in a variety of scientific fields.”
Baker is senior author of the team’s published paper in Science, which is titled “Top-down design of protein architectures with reinforcement learning,” in which the researchers concluded, “Our approach enables the top-down design of complex protein nanomaterials with desired system properties and demonstrates the power of reinforcement learning in protein design.”
Multi-subunit protein assemblies play critical roles in biology and are “the result of evolutionary selection for function of the entire assembly,” the authors wrote. As a result of evolutionary selection, they continued, “the subunits of naturally occurring protein assemblies often fit together with substantial shape complementarity to generate architectures optimal for function in a manner not achievable by current design approaches.”
Scientists carrying out de novo protein design have used a “bottom-up” hierarchical approach, starting with the monomeric structures that dock into oligomers, and working upwards to generate the final protein assemblies. This hierarchical approach does have advantages, the authors noted, and “such designed assemblies are already proving useful for biomedicine in immunobiology and other areas, as highlighted by the recent approval of a de novo–designed COVID vaccine.” However, the bottom-up approach does also have limitations. “The properties of the assembly are limited to what can be generated from the available oligomeric building blocks, at least one of the subunit-subunit interfaces must be strong enough to stabilize a cyclic oligomeric substructure in isolation, and, more generally, there is no way to directly optimize the properties of the overall assembly.”
Baker and colleagues instead looked to overcome the limitations of bottom-up protein complex design by developing a “top-down” approach that starts from a specification of the desired properties of the protein structure, such as overall symmetry and porosity, for example, and systematically builds up subunits that pack together to optimize these properties.
Reinforcement learning (RL) is a type of machine learning in which a computer program learns to make decisions by trying different actions and receiving feedback. Such an algorithm can learn to play chess, for example, by testing millions of different moves that lead to victory or defeat on the board. The program is designed to learn from these experiences and become better at making decisions over time. The authors turned to Monte Carlo tree search (MCTS), an RL algorithm that finds the optimal series of choices within a search tree. They explained, “… we turned to RL, which has achieved considerable success recently in different fields of artificial intelligence, such as self-driving cars, the AlphaGo program that defeats top human players in the game of Go, and algorithm development … We sought to develop an MCTS algorithm for generating protein complexes that builds up the monomeric subunits from protein fragments directly optimizing for prespecified global structural properties.”
To make a reinforcement learning program for protein design, the scientists—led by Isaac D. Lutz, Shunzhi Wang, PhD, and Christoffer Norn, PhD, who are all members of the Baker lab—gave the computer millions of simple starting molecules. The software then made ten thousand attempts at randomly improving each toward a predefined goal. The computer lengthened the proteins or bent them in specific ways until it learned how to contort them into desired shapes.
“Our approach is unique because we use reinforcement learning to solve the problem of creating protein shapes that fit together like pieces of a puzzle,” explained co-lead author Lutz, a doctoral student at the UW Medicine Institute for Protein Design. “This simply was not possible using prior approaches and has the potential to transform the types of molecules we can build.”
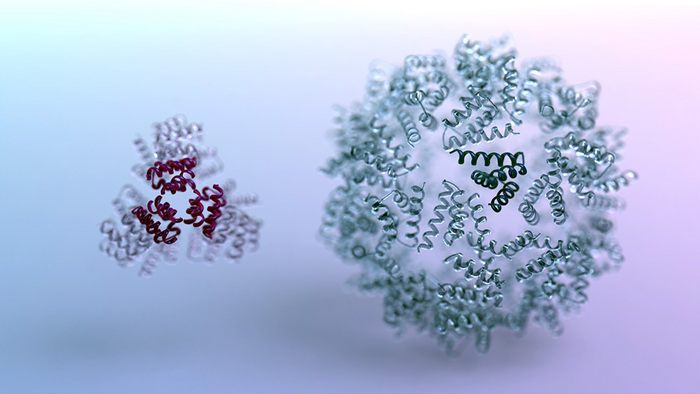
As part of their reported study, the team concentrated on designing new nano-scale structures composed of many protein molecules. This required designing both the protein components themselves and the chemical interfaces that allow the nano-structures to self-assemble. Electron microscopy confirmed that numerous AI-designed nano-structures were able to form in the lab.
The scientists manufactured hundreds of AI-designed proteins in the lab, including icosahedra and disk-shaped nanopores. Using techniques including electron microscopy, they confirmed that many of the protein shapes created by the computer were indeed realized in the lab.” Cryo–electron microscopy structures of the designed disk-shaped nanopores and ultracompact icosahedra are very close to the computational models,” they commented. “Both the icosahedra and the disk designs are distinct from any previously designed or naturally occurring structures … These structures could not have been built with previous bottom-up approaches.” Added Wang, a postdoctoral scholar at the UW Medicine Institute for Protein Design, “This approach proved not only accurate but also highly customizable. For example, we asked the software to make spherical structures with no holes, small holes, or large holes. Its potential to make all kinds of architectures has yet to be fully explored.”
As a measure of how accurate the design software had become, the scientists observed many unique nano-structures in which every atom was found to be in the intended place. In other words, the deviation between the intended and realized nano-structure was on average less than the width of a single atom. This is called atomically accurate design. “Our top-down RL approach enables the solution of design challenges inaccessible to previous bottom-up design methods,” the investigators stated.
The authors foresee a future in which this approach could enable them and others to create therapeutic proteins, vaccines, and other molecules that could not have been made using prior methods.
Researchers from the UW Medicine Institute for Stem Cell and Regenerative Medicine used primary cell models of blood vessel cells to show that the designed protein scaffolds outperformed previous versions of the technology. For example, because the receptors that help cells receive and interpret signals were clustered more densely on the more compact scaffolds, they were more effective at promoting blood vessel stability.
Co-author Hannele Ruohola-Baker, PhD, a UW School of Medicine professor of biochemistry, spoke to the implications of the investigation for regenerative medicine: “The more accurate the technology becomes, the more it opens up potential applications, including vascular treatments for diabetes, brain injuries, strokes, and other cases where blood vessels are at risk. We can also imagine more precise delivery of factors that we use to differentiate stem cells into various cell types, giving us new ways to regulate the processes of cell development and aging.”
The authors further stated, “The capability of the MCTS approach to optimize any set of specified geometric criteria in a top-down fashion provides a route to potent, multivalent cellular receptor agonists and vaccines that are custom designed to rigidly scaffold immunogen or receptor-binding monomers and precisely position them relative to one another.”