Diverse mutations can make bacterial and cancer cells resistant to chemicals, and second-generation approaches are needed. Now, a team of engineers at Penn State says it may have a way to predict which mutations will occur in people, creating an easier path to create effective pharmaceuticals.
“Structure-based drug design works very well,” said Justin Pritchard, PhD, assistant professor of biomedical engineering and holder of the Dorothy Foehr Huck and J. Lloyd Huck Early Career Entrepreneurial Professorship. “It is an amazing ecosystem of technology, but you still have to point it at a set of resistance mutations.”
Standard practice to develop drugs is to model the structure of chemicals and their cellular targets to kill specific pathogens or cancer cells. Once mutations begin to change the cells, treatment requires new drugs.
However, a variety of mutations may occur, and drug developers need to target the appropriate mutation to kill the pathogen or the cancer cells.
The researchers wanted to discover what drives which mutations to grow out in the real world so that they could choose the most effective mutations to target. In an article entitled “Multi-scale Predictions of Drug Resistance Epidemiology Identify Design Principles for Rational Drug Design,” published in Cell Reports they report that the most drug-resistant mutation was not necessarily the mutation that dominated. “Survival of the fittest” did not always hold and targeting should aim at the most probable mutation rather than the most resistant, at least for some cancers.
“We need to not just understand the biophysics,” said Pritchard. “We also need to understand the evolutionary dynamics.”
“Rationally designing drugs that last longer in the face of biological evolution is a critical objective of drug discovery. However, this goal is thwarted by the diversity and stochasticity of evolutionary trajectories that drive uncertainty in the clinic. Although biophysical models can qualitatively predict whether a mutation causes resistance, they cannot quantitatively predict the relative abundance of resistance mutations in patient populations” write the investigators in Cell Reports.
“We present stochastic, first-principle models that are parameterized on a large in vitro dataset and that accurately predict the epidemiological abundance of resistance mutations across multiple leukemia clinical trials. The ability to forecast resistance variants requires an understanding of their underlying mutation biases.” Beyond leukemia, a meta-analysis across prostate cancer, breast cancer, and gastrointestinal stromal tumors suggests that resistance evolution in the adjuvant setting is influenced by mutational bias. Our analysis establishes a principle for rational drug design: when evolution favors the most probable mutant, so should drug design.”
Drug resistance is a problem when treating diseases caused by bacteria, viruses and cancers, but the researchers chose to investigate mutations in cancers because understanding mutations in cancer cells is simpler. Mutations in bacteria and virus have two components—what happens within the cells and what happens when the bacteria or viruses spread from host to host. Because cancer is not, in humans, contagious, working with cancer cells removes a portion of the potential source of mutations.
“If we take out the community aspect of transmission, we can study just the de novo, or ‘from nothing,’ generation of mutations,” said Pritchard.
The researchers looked at existing data for leukemia and three other types of cancer. The leukemia database was the largest and most complete. They used algorithms similar to those used in modeling how chemical reactions in chemical physics take place. In this case, they used the simulations to model how evolution works.
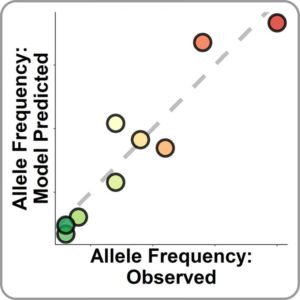
“We are trying to create a generalized approach to getting the numbers that we use in the models,” explained Pritchard. “To do this we did not ‘fit’ the model, but used data obtained from experiments and scaling.”
Creating a way to obtain data for generalized cases rather than individuals would increase the possibility of using this method for a variety of pathogens.
“We ran the model and it matched clinical data to a degree much better than I ever expected,” continued Pritchard. “We did this from first principles (basic assumptions).”
As cancer cells divide, errors that are made in the copying of DNA result in mutations. One letter of DNA might be mistakenly replaced with another, but these mistakes are not completely random. Some letters are more easily substituted for others, and so these mutations happen more often. This creates a mutation bias—some substitutions are more likely. Thus the likeliness of a mistake, and not the reduction in sensitivity to drugs, can predict the resistance mutations that real patients develop.
“We shouldn’t always focus on the strongest resistance mutation because there are other evolutionary forces that dictate what happens in the real world,” according to Pritchard. “Sometimes drug resistance relies on biased random events.”
The researchers found that biased random mutations played a big part in the evolution of resistance in leukemia. They found similar results with breast, prostate and stomach cancers, although the effect size was not as large.
“The data are not quite as strong in the prostate and breast cancer setting,” said Pritchard. “In non-small cell lung cancer we didn’t see this effect at all.”
According to the researchers, there are lots of places where evolutionary bias creates an abundance of mutations that are not the most resistant strains, but it is a spectrum with leukemias on one end; breast, prostate and stomach cancers in the middle; and non-small cell lung cancer on the other end.
“Our analysis establishes a principle for rational drug design: When evolution favors the most probable mutant, so should drug design,” noted the research team.






