CRISPR pioneer Jennifer Doudna, Ph.D., has co-founded a new company, Mammoth Biosciences, that aims to democratize gene-editing technology by developing a platform that the company and its partners may use to launch an “endless variety of tests for biosensing in healthcare, as well as across industries such as agriculture, manufacturing, forensics, and more.” Belying its ponderous name, Mammoth chooses to emphasize the speed with which its technology may lead to new applications, which will, themselves, speed test results to patients in clinical settings, or people at home.
According to Mammoth, users won’t have to take multiple trips to the clinic and wait days for their results. Instead, they can “add a sample,” “take a picture,” and “get results” in 30 minutes.
“Our platform has two form factors to enable its use in many different settings,” Mammoth’s website indicates. “For use in the field and at home, we use a simple paper-based test. For this system, the sample is applied to the detection card, resulting in a visible color change. To achieve the maximum sensitivity and accuracy in real time, we can leverage a smartphone to detect the signal in liquid containing the sample of interest.”
Mammoth is licensing technology from the University of California, Berkeley, where Doudna, a professor of molecular and cell biology and of chemistry, leads research into CRISPR (clustered regularly interspaced short palindromic repeats). Of particular relevance is a CRISPR technology developed by Doudna and colleagues called DETECTR, which stands for DNA endonuclease targeted CRISPR trans reporter.
Announced last February as “a simple method that may lead to fast, reliable medical tests,” DETECTR has also been featured in a recent paper (“CRISPR-Cas12a Target Binding Unleashes Indiscriminate Single-Stranded DNase Activity”) in the journal Science.
“By combining Cas12a ssDNase [single-stranded DNase] activation with isothermal amplification, we create a method termed DNA endonuclease-targeted CRISPR trans reporter (DETECTR), which achieves attomolar sensitivity for DNA detection,” wrote the article’s authors. “DETECTR enables rapid and specific detection of human papillomavirus in patient samples, thereby providing a simple platform for molecular diagnostics.”
As this statement indicates, DETECTR makes use of an alternative CRISPR nuclease. Most CRISPR development has centered on the Cas9 nuclease. Alternative nucleases, such as Cas12, are “programmable,” like Cas9, because they may be paired with different guide RNAs, which direct DNA-cutting nucleases to their targets. Working with different nucleases may prove fruitful, however, because they present different capabilities.
“Our initial platform makes use of two unique CRISPR proteins,” details the Mammoth website. “The first, Cas12, binds and cuts DNA, the material that makes up the genomes of all living things on Earth. The second, Cas13, recognizes and cuts a related molecule called RNA. Together, these two proteins enable us to sense virtually any type of nucleic acid.”
Like its molecular cousin Cas9, Cas12a snips DNA. But instead of snipping only the DNA strand it binds, Cas12a chops other DNA, too. “We started to see this surprising activity where it would just start cutting random stuff,” said Lucas Harrington, a graduate student in Doudna's lab and a coauthor of the Science paper.
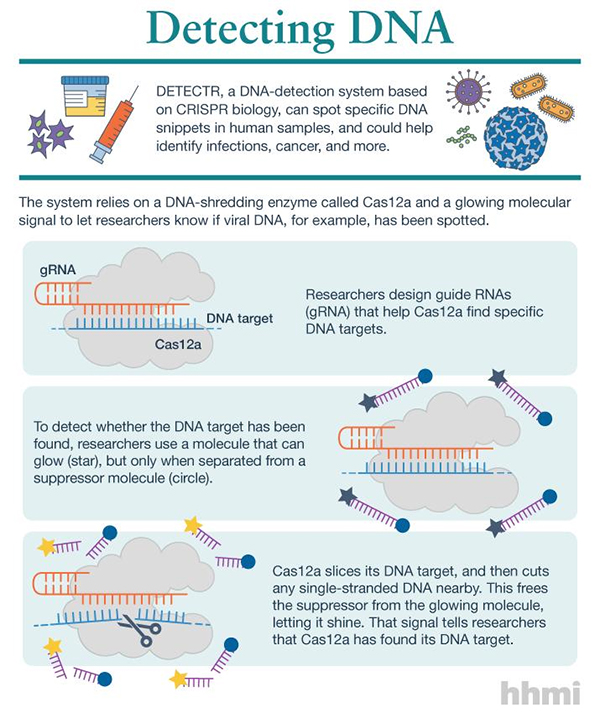
Under certain circumstances, the enzyme turns into a DNA-shredding machine, slicing up any single-stranded DNA nearby, the researchers saw. But this wasn't indiscriminate destruction. For the machete action to begin, Cas12a first has to find a precise DNA target. Researchers can program that target by adding a guide RNA molecule to tell Cas12a what to look for
Once Cas12a locks onto and snips the target, it then begins shredding all of the single-stranded DNA it can find. But for the system to be useful, Doudna and colleagues needed a way to see when Cas12a starts this molecular mayhem, signaling that it has found its target. So the researchers used a glowing molecule—an easy-to-spot flare—linked by a single strand of DNA to a suppressor molecule that prevents the glow. When Cas12a turns into a machete, it slices the DNA strand that links these two molecules together. This removes the suppressor, letting the glowing molecule shine—a signal researchers can detect.
The team then put their DNA detective to the test. Working with Joel Palefsky, M.D., and his team at the University of California, San Francisco, the investigators hunted for DNA signals from two types of cancer-causing human papillomavirus (HPV): type 16 and type 18. Researchers obtained 25 DNA samples taken from people who had no HPV infection, one type of virus, or both types. For HPV16, DETECTR made the right call for all 25 of the samples. For HPV18, DETECTR got it right for 23 of 25 samples. The ones it missed gave weak signals that can likely be improved with different guide RNA design, Doudna noted.
Compared with current methods to detect HPV, DETECTR is “simpler, quicker, and does not require specialized equipment,” Harrington asserted. That could make the system useful in resource-limited health clinics and for point-of-care diagnostics.
Doudna's team's method could easily be applied to other types of viral or bacterial infections, and even cancer markers, chromosomal abnormalities, or other genetic signals, Harrington commented at the time the study appeared.
In addition to the protein and guide RNA components, the Mammoth technology also includes a reporter molecule. Once Cas12 or Cas13 finds its matching target, the protein breaks apart the reporter molecule, which produces a color change. This signal indicates the presence of a specific DNA or RNA sequence.