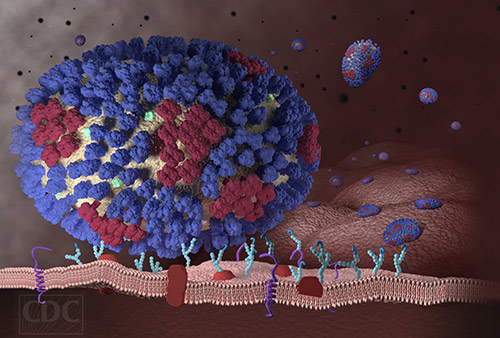
To turn back the invading force that is influenza, drugs can directly block the virus or adopt an indirect approach, a kind of scorched earth strategy. With the indirect approach, drugs could be used to prevent the virus from taking over host capabilities. Doing so is difficult, however, because influenza-host interactions are poorly mapped.
Enter “big data.” It is being used to survey the biochemical landscape of influenza-host interactions. Already, it has identified 20 previously unrecognized host proteins that are required for influenza A virus (IAV) interactions. One host protein in particular has been shown to be a particularly promising drug target. This protein, called UBR4, is useful to the virus because of its role in supporting viral budding from the host cell membrane.
These results were generated by scientists based at the Sanford Burnham Prebys Medical Discovery Institute (SBP) and the Icahn School of Medicine at Mount Sinai. These scientists, led by SBP’s Sumit Chanda, Ph.D., published their work December 9 in the journal Cell Host and Microbe, in an article entitled, “Meta- and Orthogonal Integration of Influenza “OMICs” Data Defines a Role for UBR4 in Virus Budding.”
Denying influenza access to host resources is like preventing an invader from “living off the land.” This approach, the scientists reported, could prove to be even more effective than trying to directly block the virus.
“Traditionally, physicians have treated the flu with drugs that directly block the influenza virus,” said Dr. Chanda. “Although these drugs have been helpful, many patients fail to respond because viruses, especially IAV, can mutate, rendering them resistant to available drugs. Our research efforts are focused on finding unalterable host molecules—the ones within our bodies—that viruses hijack to spread and create full blown infections.”
Influenza viruses cannot replicate on their own. They can only carry a few genes—about a dozen or so—compared to a human genome comprising more than 20,000 genes. To ensure their survival, flu viruses rely on co-opting molecular machines in the infected host, which they use to their advantage to grow and spread.
Details of the big data approach were given in the Cell Host and Microbe article: “[We] performed a meta-analysis of data from eight published RNAi screens and integrated these data with three protein interaction datasets, including one generated within the context of this study. Further integration of these data with global virus-host interaction analyses revealed a functionally validated biochemical landscape of the influenza-host interface, which can be queried through a simplified and customizable web portal.”
The study also showed that blocking UBR4 in human cells (in vitro) and mice (in vivo) reduced IAV replication and pathogenesis, establishing proof-of-concept of the strategy to target UBR4 as an influenza treatment. “The putative ubiquitin ligase UBR4,” the article noted, “associates with the viral M2 protein and promotes apical transport of viral proteins.”
The article concluded that “the integrative analysis of influenza OMICs datasets illuminates a viral-host network of high-confidence human proteins that are essential for influenza A virus replication.”
“We anticipate that the approach described in this study, which is packaged as an accessible web interface, will provide a bridge for those on the frontlines of biomedical discovery and therapeutic development to leverage 'big data' and achieve transformative treatments for unmet medical needs, added Dr. Chanda”