“Round up the usual suspects!” This expression, which bespeaks expediency, could apply to the gene editing tools known as cytosine base editors (CBEs). CBEs, which are supposed to convert selected cytosines (Cs) in the human genome to thymines (Ts), may fail to distinguish between Cs in need of conversion, and nearby Cs that should be left alone. If any of the latter Cs, or “bystander” Cs, were anxious about being rounded up, they can now heave a sigh of relief. There is a new CBE in town, and it can chase down targeted Cs while giving bystander Cs a pass.
The new CBE, which was engineered at Rice University, can increase the accuracy of CRISPR-based edits in disease sequence models up to 6000-fold compared with the BE4max base editor, which is currently considered state of the art. “I believe our technology’s level of precision is going to be a significant contributor toward treating genetic disease,” said Xue Sherry Gao, PhD, the Rice University biomolecular engineer who led the development of the new CBE.
Gao and colleagues described their work in an article, “Single C-to-T substitution using engineered APOBEC3G-nCas9 base editors with minimum genome- and transcriptome-wide off-target effects,” that appeared July 15 in Science Advances.
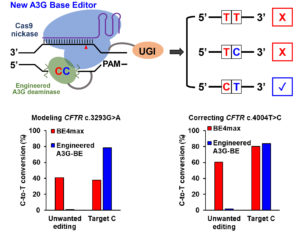
“To improve the precision of CBE, we identified and engineered the human APOBEC3G (A3G) deaminase,” the article’s authors wrote. “When fused to the Cas9 nickase, the resulting A3G-BEs exhibit selective editing of the second C in the 5′-CC-3′ motif in human cells.”
In the human genome, which consists of three billion Cs, Ts, As (adenines), and Gs (guanines), even one incorrect base—a mutation—can lead to genetic diseases. “T-to-C mutations called single nucleotide polymorphisms account for somewhere around 38% of human pathogenic diseases,” Gao said. “Cytosine base editors provide great promise to potentially treat these diseases by reversing the C mutation back to T.”
“However, when there is a ‘bystander’ C located right upstream of the targeted C, the previous technology could not distinguish between the Cs, and both would be changed to Ts,” she added. “We really only want to correct the disease-relevant C to a T and leave the bystander C unmodified.
“That provided the motivation for this project. We want to engineer a new cytosine base editor that can precisely modify the single targeted C while minimizing the unwanted C editing when consecutive ‘CCs’ are positioned in the editing window.”
The Gao lab seeks to develop base editors through a series of protein-engineering efforts. The new CBEs, called A3G base editors, have dramatically increased precision by only editing the second of consecutive Cs.
To put their tests in “disease-relevant contexts,” the Gao lab used their tools to modify human cells to create cystic fibrosis and several other disease model cell lines. All showed significant success at precisely creating the desired pathogenic C-to-T mutation, particularly the cystic fibrosis cells, which all three of the A3G-BE variants perfectly modified more than 50% of the time compared to 0.6% for BE4max.
The Gao lab also tested its new A3G-BEs’ potential to correct mutations in disease treatment applications, including cystic fibrosis, holocarboxylase synthetase deficiency, and pyropoikilocytosis, a type of anemia.
In experiments on cell models containing pathogenetic mutations, A3G-BEs significantly outperformed BE4max. In the case of holocarboxylase synthetase deficiency, the editor perfectly corrected only the target C nucleotides in more than 50% of the sequences, with a 6496-fold higher correction than BE4max.
“We also identified 540 human pathogenic single nucleotide polymorphisms that could be precisely correctable by our A3G-BEs,” Gao said. “A3G-BE also appears to decrease off-target edits (unwanted edits to other parts of the genome that could introduce mutations) at both the DNA and RNA levels.”