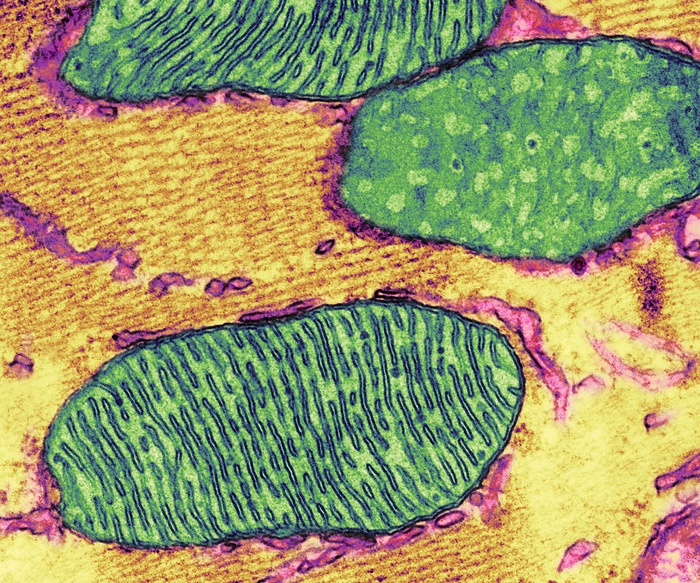
It has been known that mitochondrial play a crucial role in the metabolism and energy production of cancer cells. However, the relationship between the structural organization of mitochondrial networks and their functional bioenergetic activity at the level of whole tumors is not fully understood. Now, researchers from the UCLA Jonsson Comprehensive Cancer Center used positron emission tomography (PET) in combination with electron microscopy to generate three-dimensional ultra-resolution maps of mitochondrial networks in lung tumors of genetically engineered mice.
Their findings are published in Nature in an article titled, “Spatial mapping of mitochondrial networks and bioenergetics in lung cancer,” and may lead to new approaches to cancer treatment.
The researchers categorized the tumors based on mitochondrial activity and other factors through deep learning, quantifying the mitochondrial architecture across hundreds of cells and thousands of mitochondria throughout the tumor.
Two main subtypes of non-small cell lung cancer (NSCLC)—adenocarcinomas and squamous-cell carcinomas—were examined and distinct subpopulations of mitochondrial networks within these tumors were found.
The researchers discovered that the mitochondria organize themselves with organelles such as lipid droplets to create unique subcellular structures that support tumor cell metabolism and mitochondrial activity.
“Our study represents a first step towards generating highly detailed three-dimensional maps of lung tumors using genetically engineered mouse models,” said David Shackelford, PhD, associate professor, medicine, University of California, Los Angeles. “Using these maps, we have begun to create a structural and functional atlas of lung tumors, which has provided us valuable insight into how tumor cells structurally organize their cellular architecture in response to the high metabolic demands of tumor growth. Our findings hold promise to inform and improve current treatment strategies while illuminating new directions from which to target lung cancer.”
“Our study has uncovered a novel finding in the metabolic flux of lung tumors, revealing that their nutrient preference may be determined by the compartmentalization of their mitochondria with other organelles, either relying on glucose (“sugar”) or free fatty acids (“fat”),” said Mingqi Han, PhD, a postdoc in Shackelford’s lab. “This discovery has important implications for developing effective anticancer therapies that target tumor-specific nutrient preferences. Our multi-modality imaging approach has enabled us to uncover this previously unknown aspect of cancer metabolism, and we believe that it can be applied to other types of cancer, paving the way for further research in this area.”