February 15, 2013 (Vol. 33, No. 4)
Thalyana Smith-Vikos
miRNAs have emerged as important fine-tune regulators of a large host of genes that play a role in many cellular processes and signaling pathways.
These molecules function in a variety of diseases, such as cancer and inflammation, highlighting their potential as therapeutics and diagnostic biomarker tools.
CHI’s upcoming “microRNA: Targets and Tools for Therapeutic Development” conference will address the latest discoveries regarding miRNA-mediated disease regulation and introduce new methodologies for conducting these studies in a laboratory or clinical setting.
Much of the latest research in miRNA profiling for biomarker analysis has focused on improving methods of miRNA detection. Daniel Pregibon, Ph.D., CTO of Firefly BioWorks, will discuss the firm’s latest technology, FirePlex™, a method of miRNA detection that has been used by both academic and clinical researchers.
Dr. Pregibon explains that there are two main classes of technologies widely available for miRNA profiling studies, either using microarrays or deep sequencing to profile all miRNAs in a single sample, or using single assays to examine individual miRNA expression across many samples. Unfortunately, these approaches are not suitable for performing biomarker validation studies, which are fundamental in the development of diagnostic tests.
“What we’re seeing is that there was really nothing in that middle range, if you wanted to look at 25–50 miRNAs over hundreds or thousands of different samples,” he says. Thus, FirePlex was designed to help researchers with high-throughput validation of a certain subset of miRNAs of particular interest for their studies.
In fact, because each researcher may be interested in looking at a unique set of miRNAs, Dr. Pregibon reported that 99% of the panels they have made are custom panels, which can be quickly prepared within one week. Additionally, FirePlex panels have been developed to run on benchtop flow cytometers, so additional equipment does not need to be purchased to run an assay.
The FirePlex technology is built on encoded hydrogel particles, instead of using a glass surface, as would be utilized in microarrays. Dr. Pregibon explains that using a hydrogel instead of a glass surface allows each molecule to have many more degrees of freedom, which is beneficial from a thermodynamics standpoint. This hydrogel base for capturing miRNAs allows for better sensitivity and specificity. Polyethylene glycol (PEG) is used as a substrate to eliminate nonspecific binding of proteins, lipids, or other complexes.
FirePlex can also be used with crude samples because the assay is based on post-hybridization labeling (the miRNAs are labeled after being hybridized throughout the hydrogel volume), so any debris will be washed away after capturing the miRNAs. Dr. Pregibon notes that minimal manipulation of the sample also reduces any bias that is introduced as a result of the RNA isolation method used, which can lead to variable RNA yield or selective enrichment. FirePlex has also been expanded to profile miRNAs in cell lysates, fresh tissue, FFPE, and serum/plasma.
SomaGenics has developed a method, miR-IDirect, for direct detection of miRNAs from a plasma sample without requiring total RNA isolation. Sumedha Jayasena, Ph.D., vp of technology and therapeutic development, says the miR-IDirect platform, which detects circulating miRNA in plasma, incorporates SomaGenics’ qPCR-based miR-ID technology.
The miR-ID method provides quantification of small RNAs by first circularizing them, then generating cDNA by rolling circle amplification of the miRNA circles, and finally further amplifying the cDNA by qPCR using 5´-overlapping PCR primers. miR-ID has previously been applied to detecting miRNAs in total cellular RNA as well as purified from various biological fluids.
Validated miR-ID assays have been developed for about 100 different miRNAs to date, with more in development. The miR-ID technology can discriminate miRNAs that carry terminal modifications, such as 2´-OMe groups at 3´-ends, according to Dr. Jayasena.
He says that the miR-IDirect technology was developed to overcome certain limitations that have been encountered in analyzing circulating miRNAs as potential biomarkers. He explains that the latest wave of miRNA-based biomarker research has been primarily focused on their identification in biological fluids, especially in blood, as it was discovered that miRNAs are surprisingly stable in biofluids, and their expression profiles often correlate with particular disease states.
Current methods require total RNA or miRNA isolation, which can lead to bias against certain miRNAs and insufficient sensitivity to detect those miRNAs that are present at low levels. Further, a normalization standard, based on an endogenous miRNA, has not been established for profiling circulating miRNAs. A control miRNA can be spiked into samples, but because (unlike endogenous miRNAs) it is unprotected from attack by plasma nucleases, it is likely to be degraded to an unpredictable extent.
miR-IDirect addresses these issues, providing more accurate miRNA detection and quantification. The method “should facilitate research on circulating miRNAs, and, with minor adaptations, miRNAs in other biological fluids besides blood,” Dr. Jayasena says. “Our technology will help eliminate variability associated with current methods, while improving sensitivity in miRNA quantification and accelerate clinical validation,” Dr. Jayasena states.
Evaluating miRNAs in Circulation
Dominik M. Duelli, Ph.D., assistant professor at Rosalind Franklin University of Medicine and Science, has also been interested in circulating miRNAs, and his lab has evaluated the most accurate and reproducible methods for miRNA detection and quantification from plasma via qRT-PCR.
Dr. Duelli explains that the idea behind testing these various parameters of miRNA profiling stemmed from inconsistencies in detecting certain miRNAs in plasma. “When we first started our work, the original idea for a biomarker measured in blood plasma was that it should reflect what’s inside the malignancy, so an miRNA that’s high inside the cell should also be high outside the cell,” Dr. Duelli explains.
“We profiled miRNAs in an array of cell-line cultures and saw that 60–70% of the miRNAs had the same profile outside and inside cells. But some miRNAs appeared to be exclusively released 90% or more out of the cell. Another category was miRNAs that were retained and were not released at all.” Dr. Duelli wanted to know if this was in fact a biological phenomenon, or were the miRNAs simply not detectable based on the experimental methodology?
His lab began by measuring miR-16 (highly abundant in plasma) and miR-223 (low abundance in plasma) in fresh plasma from 16 individuals to determine the optimal parameters for miRNA profiling. They first noticed that anticoagulants like citrate or KOx/NaF are a much better option than heparin for miRNA quantitation. Heparin can inhibit RT and polymerases, and miRNA detection was possible only if they diluted the heparin or treated raw plasma with heparinase.
Next, Dr. Duelli and his lab evaluated different methods for RNA isolation and noticed that some reagents can selectively precipitate certain miRNAs, so that some miRNAs may not be detected at all in a sample. Instead, using a silica membrane or beads for RNA extraction prevents polymerase inhibitors from co-purifying with miRNAs and leads to better purity and yield.
Additionally, Dr. Duelli notes that using a Taq polymerase such as Hemo KlenTaq™ (New England Biolabs) can solve this problem because some major inhibitors cannot bind to this truncated polymerase. Dr. Duelli’s lab observed a 30-fold increase in miR-16 and miR-223 expression simply by including Hemo KlenTaq in the reaction.
Dr. Duelli’s lab has also used fluorescent SmartFlare™ RNA Detection Probes (EMD Millipore), which utilize a nonenzymatic approach for rapid direct miRNA quantification in live samples. They were able to measure miR-16 expression in plasma within minutes to one hour after venipuncture, he says.
Dr. Duelli expects this technology to be highly useful in the clinical setting. “You can measure miRNA levels right after chemotherapy and correlate it with tumor progression; if the tumor is shrinking, you can measure if these miRNAs go away. It comes at a cost to sensitivity because there is no amplification, but it can be used in plasma or other samples.”
Viral-Mediated miRNA Therapeutics
Along with investigating miRNAs as biomarkers and diagnostic tools, there have also been a number of advances in the field of miRNA therapeutics. Benjamin tenOever, Ph.D., Fishberg professor of medicine at Mount Sinai School of Medicine, has been investigating how miRNAs can be utilized in engineered RNA-based vectors.
Dr. tenOever’s lab has developed a method for exploiting endogenous miRNAs to regulate tissue tropism of viral vectors. “Because viruses lack a mechanism for antagonizing miRNA function, we can exploit miRNA expression to control viruses,” he says. “We have shown that if you incorporate targets for a cell-specific miRNA into an RNA virus genome, you can create a virus that looks identical at a protein level but would be selectively blocked from infecting these particular cells.”
The other side of Dr. tenOever’s research focuses on engineering viral vectors to produce functional miRNAs, which can be used as a therapeutic platform to deliver miRNAs (or other small RNAs) to any tissue in the body. Dr. tenOever states that, while it was known that RNA viruses do not produce miRNAs, it was unclear whether viruses lack the capacity to do so, or whether this activity was perhaps detrimental to the viral life cycle.
His lab addressed this question by incorporating primary miRNAs (pri-miRNA) into various RNA viral vectors that localize to the nucleus or the cytoplasm in different tissues. In all cases, mature miRNAs were properly processed and loaded into the RISC complex. In fact, with cytoplasmic viruses, Drosha, an RNase, was exported out of the nucleus to process the artificial pri-miRNA.
It appears that most RNA viruses are capable of producing functional miRNAs, but do not do so naturally, presumably to prevent some degree of self-attenuation. “What we can capitalize on now is to convert an miRNA that behaves like a tailor-made, sequence-specific siRNA. We can then adapt any RNA virus, regardless of tropism, and use it to generate siRNAs to silence a desired host gene,” Dr. tenOever reported.
The RNA viruses only produce miRNAs for 7–10 days, but transient cytoplasmic RNA viruses have become very useful in Dr. tenOever’s lab for a variety of applications including the reprogramming of fibroblast cells into iPSCs by introducing a few miRNAs.
“The advantage of our system is that we are not going into the nucleus with our vectors and we’re not integrating into the genome; we’re only there for 7–10 days and then our vectors disappear, but by then we have reprogrammed our cells to become pluripotent stem cells.”
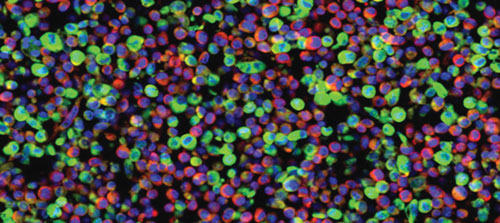
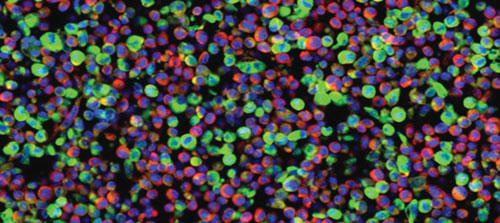
Cellular delivery of miRNAs via cytoplasmic-based vectors: Colors depict the nucleus (blue), cytoplasm (red), and the virus-derived artificial miRNA (green). This biological activity can be harnessed as a means of delivering siRNAs to a tissue of interest. [Mount Sinai School of Medicine]