February 1, 2012 (Vol. 32, No. 3)
James C. Samuelson, Ph.D. senior scientist New England Biolabs
Thomas B. Causey, Ph.D. fermentation process manager New England Biolabs
Mehmet Berkmen, Ph.D. New England Biolabs.
Involvement of Disulfide Bond Isomerase Improves Protein Folding
Disulfide bonds are an essential stabilizing feature of many proteins. More than 50% of human ER proteins are estimated to contain disulfide bonds (dsb) and the majority of secreted proteins also contain dsbs.
Using bacteria to produce these proteins efficiently presents a significant challenge because the correct pairing of cysteines in a protein with multiple disulfide bonds is inherently fraught with error. Misoxidation of the incorrect pairs of cysteines results in misfolding and low yields.
Nature has resolved this issue by shuffling the incorrect disulfide bonds in misfolded protein into their native correct pairing by the activity of disulfide bond isomerases. Protein disulfide isomerase (PDI) carries out dsb oxidation and isomerization in the ER of all eukaryotes.
In many Gram-negative bacteria (including E. coli) the cooperative action of DsbA and DsbC oxidizes proteins within the periplasmic space. This is also true within commonly used E. coli protein-expression strains, with the exception of engineered strains such as the Origami™ strains from EMD Biosciences and the SHuffle® strains from New England Biolabs. These commercial strains have been similarly engineered to possess an oxidative cytoplasmic environment that favors disulfide bond formation. The SHuffle strains have been further modified to support robust production of dsb-containing proteins.
Dsb Formation in the SHuffle Strains
SHuffle strains contain a unique trio of modifications that produce robust disulfide bond formation in the cytoplasm.
First, the cytoplasmic environment is altered by elimination of glutathione reductase (gor gene product) and thioredoxin reductase (trxB gene product). Since the combination of gor and trxB deletions are lethal, a suppressor mutation in the ahpC gene is necessary for the SHuffle (and Origami) strains to maintain viability.
Second, SHuffle strains are uniquely engineered to overexpress DsbC within the cytoplasm. The DsbC enzyme acts as a disulfide bond isomerase and “shuffles” mis-oxidized cysteine pairs allowing the recombinant target protein to achieve its properly folded confirmation. Due to the action of DsbC in the SHuffle cell, less target protein proceeds down the paths of protease degradation or inclusion body formation.
Finally, the SHuffle strains were engineered from robust parent strains capable of tightly controlled protein expression. For example, SHuffle T7 Express (B strain) and SHuffle T7 (K-12 strain) both express the T7 RNA polymerase from the lac operon, whereas most other T7 expression strains utilize the DE3 prophage.
T7 expression in DE3 strains is known to be somewhat uncontrolled and this can have very detrimental effects on cell growth and protein yield when the target protein is even mildly toxic, which is a common occurrence with heterologous dsb-containing proteins. If strictly controlled T7 expression is required, then lysY strains are also available.
The lysY gene product is a variant of T7 lysozyme containing a mutation that eliminates “lysozyme” function on the E. coli cell wall, but the ability of LysY to inhibit T7 RNA polymerase function is unaffected. During the pre-induction phase, a constant low level of LysY inhibitor protein is produced to inactivate any basal expression of T7 RNA polymerase.
Fermentation Performance
SHuffle strains can be easily grown to high cell densities, OD600 25–50, in batch culture using rich growth medium. Although process optimization for production of one’s specific protein will be required there are some general principles to follow when beginning the optimization.
A general batch fermentation medium that can be used with SHuffle is: 2% soytone, 1% yeast extract, 2% glycerol, 37 mM KH2PO4, 120 mM Na2HPO4, 0.5 mM K2SO4, and 5 mM MgCl2. The pH of the medium should be adjusted to 7.0 with 30% NH4OH.
Glycerol is typically used as a carbon source when producing recombinant proteins in E. coli to achieve high cell densities. Typically, E. coli production strains grow well in media containing 5–10% glycerol.
However, our experience indicates that SHuffle does not grow well in high glycerol concentrations. SHuffle strains grow optimally when the glycerol concentration is below 2%. Accordingly, to achieve high cell densities multiple bolus doses of glycerol may be added so long as the final glycerol concentration in the fermentor is not greater than 2%. One bolus dose at the induction point is usually sufficient.
Other carbon sources such as glucose can be used but organic acid accumulation and inhibition of growth may occur when the cells are grown at low dissolved oxygen.
Other factors that affect recombinant protein yield in SHuffle include inducer concentration, induction temperature, and dissolved oxygen. A standard fermentation process involves pre-induction growth at 30°C followed by induction of protein expression at 16–20°C for 16–20 hours. Many of the recombinant proteins produced in SHuffle are more active when the dissolved oxygen is controlled between 5–10% of air saturation.
We also find it is sometimes beneficial to use a nonsaturating concentration of inducer to obtain correctly folded, soluble protein. For common promoters such a Ptac, Ptrc, Plac, and PT7, IPTG concentrations in the range of 20–100 µM may result in the greatest yield of soluble, active protein. This recommendation is especially relevant when expressing a target protein fused to maltose binding protein since the pMALc vectors employ a strong tac promoter.
A standard 10 L SHuffle fermentation scheme is depicted in Figure 1.
Preferably a single colony from a fresh transformation—a colony struck from a glycerol stock may also be used—is inoculated into 500 mL of LB broth. The culture is shaken at 30°C and 300 rpm for 12–16 hours until the OD600 reaches two. Then, the appropriate volume of seed culture is added to the fermentor to obtain an initial OD600 of 0.01.
The fermentor temperature is maintained at 30°C, pH is controlled at 7.0 by automatic addition of 30% NH4OH and 23 NH3PO4, and the dissolved oxygen level is maintained at 10% of air saturation.
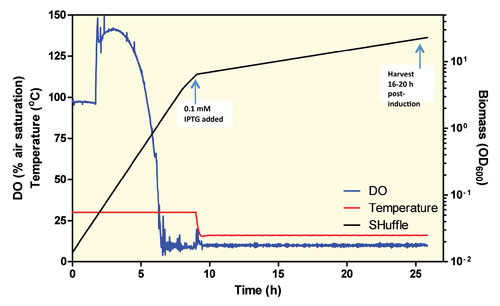
Once the culture reaches 10 OD600 it is ready to induce (Figure 2). Typically the temperature is reduced to 16°C, IPTG is added to 0.1 mM, and the induced culture is allowed to grow for an additional 18 hours.
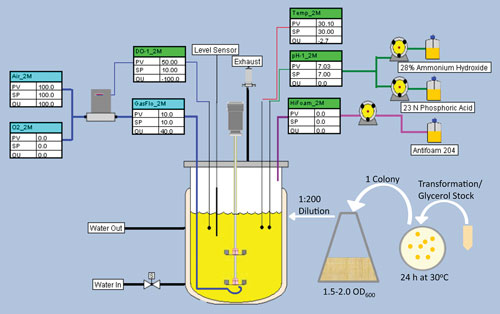
Figure 1. Standard SHuffle fermentation scheme: The fermentation was performed using a New Brunswick Scientific BioFlo 415 fermentor. Parts of the figure were generated with NBS Biocommand software.
Applications
SHuffle strains yield proteins with complex arrays of disulfide bonds. These E. coli strains are ideally suited for rapid screening of targets that may eventually be produced in higher organisms such as yeast or CHO cells.
Commercial protein production using a high density fermentation process can also be achieved with SHuffle strains. For example, we have achieved purification yields of 3.2 grams of recombinant target protein per 562 grams of cells (wet weight) from a single 10 L fermentation run.
Other examples emerging from our studies and the work of collaborators highlight the potential of the SHuffle strains—full-length functional IgG molecules and single-chain polypeptides with up to 35 cysteines (17 possible dsbs) have been produced. In simpler cases, proteins with only one disulfide bond may also be produced at higher levels if cytoplasmic expression is employed.
Most proteins with multiple disulfide bonds require the activity of a disulfide bond isomerase in order to reach their native folded state. Therefore, the availability of DsbC within the SHuffle cytoplasm sets these strains apart from other first-generation trxB, gor protein-expression hosts.
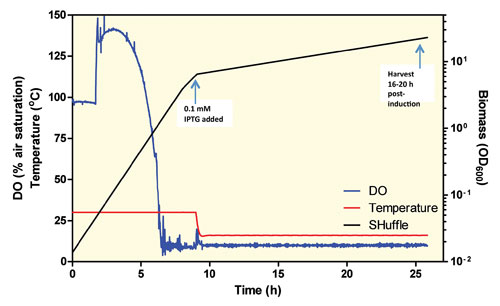
Figure 2. 10 L SHuffle Fermentation: The dissolved oxygen increases because 8 psi is applied to the tank to minimize the amount of oxygen enrichment necessary when the cell density is high. IPTG is added at 9 hours ~10 OD600 and the temperature is decreased to 16°C.
James C. Samuelson, Ph.D., is a senior scientist, Thomas B. Causey, Ph.D., is a fermentation process manager, and Mehmet Berkmen, Ph.D. ([email protected]), is a staff scientist at New England Biolabs. Web: www.neb.com.