In microRNA (miRNA) profiling, researchers confront a good news / bad news scenario. The good news is: miRNA profiles show promise in biomarker discovery. The bad news is: miRNA profiles generated by different research teams don’t always overlap—at least not so much as to inspire confidence that researchers are converging on clinically useful assays.
Part of the problem is that miRNA molecules consist of nucleotide sequences that are so short they pose challenges with respect to probe design and label selection. Different miRNAs may have widely varying melting temperatures for annealing reactions, they may be present in concentrations that vary by orders of magnitude, and they may be hard to distinguish from miRNA precursors and variants arising from post-transcriptional modifications. Consequently, identifying and quantifying miRNA molecules is inherently challenging, even though they are relatively stable compared to messenger RNA (mRNA) molecules.
Another part of the problem is that miRNAs are found in different contexts—in tissues, in isolated cells, in extracellular vesicles, and bound with proteins while swimming in biofluids. Different miRNA sample types call for different kinds of extraction techniques, and even extraction techniques of the same kind may vary in small but ultimately significant ways, leading to divergent results. Other sources of variability include miRNA measurement and data interpretation.
To overcome variability, and thus find much desired common ground, researchers and tool providers alike are working hard to standardize miRNA profiling protocols, reconcile findings from disparate research teams, and develop applications that deliver reliable diagnostic and prognostic findings. As they advance their work, researchers and tool providers fulfill, step by step, the potential for miRNA biomarkers that has been demonstrated in many recent studies.
MicroRNAs as Biomarkers
Many studies have shown how miRNA profiles may differ depending on health status. This point is easily confirmed. All the reader need do is open a search engine and plug in a few key terms: miRNA, biomarker, review, and cancer—or another condition, such as cardiovascular disease or neurodegenerative disease.
Besides looking back, readers may survey current reports on miRNA biomarker research. For example, this past year, new findings have been announced for the following conditions:
- Bronchopulmonary dysplasia, a disease that can lead to death or long-term disease in low-birth-weight infants (University of Alabama at Birmingham, JCI Insight).
- Atrial fibrillation, an irregular heart rhythm often overlooked because of a lack of symptoms (Tokyo Medical and Dental University, Circulation).
- Lou Gehrig’s disease, or amyotrophic lateral sclerosis (ALS), a neurodegenerative disease that destroys
nerve cells and causes permanent disability (Tel Aviv University, The Journal of Neuroscience). - Cognitive performance deficits related to sleep deprivation. (University of Pennsylvania School of Medicine, SLEEP 2018).
- Alzheimer’s disease (Indiana University, Nature Scientific Reports).
- Sepsis, an extreme and life-threatening reaction to infection that can trigger inflammation throughout the body (Columbia University Irving Medical Center, Nature).
- Blood doping, the transfusion of autologous red blood cells to enhance athletic performance (Duke University, British Journal of Haematology).
MicroRNA Profiling Essentials
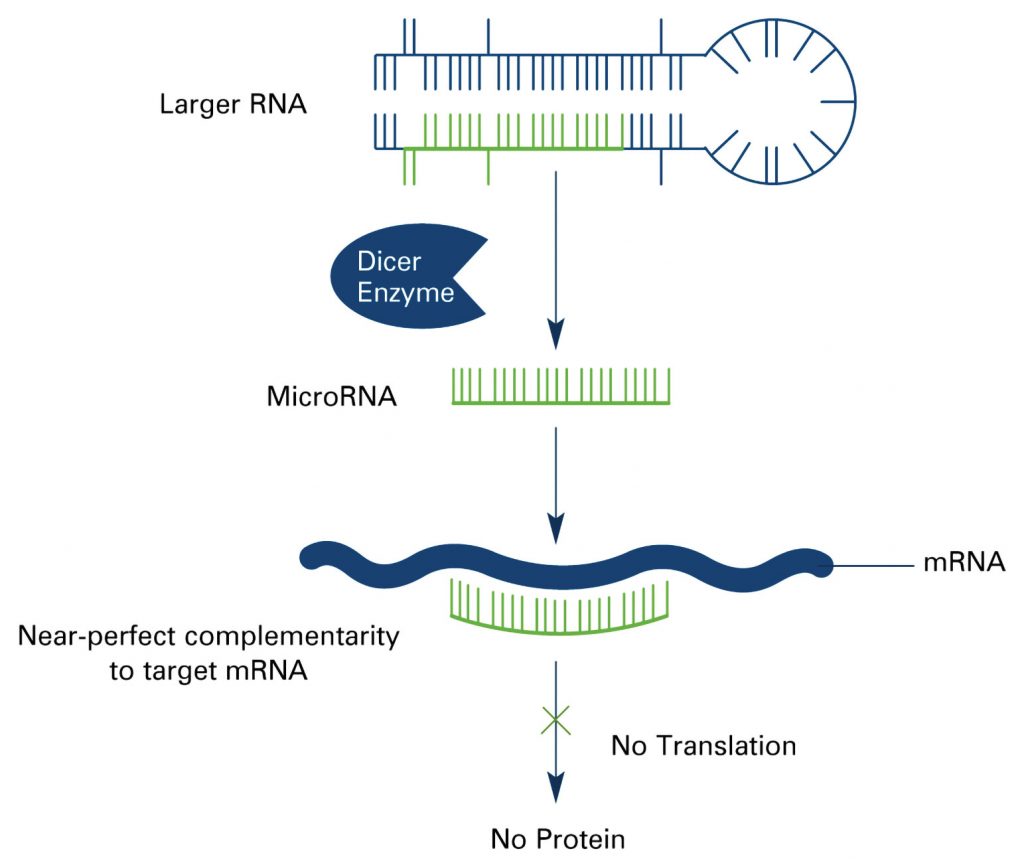
MicroRNAs, noncoding RNAs that are about 22 nucleotides long, regulate gene expression by binding to mRNAs, inhibiting their translation or resulting in their degradation. Although miRNAs are not especially numerous—only about 1000 are thought to be derived from the human genome—each miRNA may regulate hundreds of mRNAs. Consequently, miRNAs participate in many biological processes, including cell differentiation, proliferation, and apoptosis.
When miRNA expression goes awry, the consequences may include the dysregulation of gene expression, which in turn may result in disease. Fortunately, miRNAs are relatively stable in various sample types—in liquid biopsy samples as well as fresh and fixed tissue samples—raising the possibility that miRNAs can be sufficiently well quantitated to disease signatures to be recognized.
MicroRNA quantitation and profiling essentials may be studied from many sources, including a magisterial article that appeared in Nature Reviews Genetics. This article was prepared by a team of researchers based at the Fred Hutchinson Cancer Research Center and led by Muneesh Tewari, M.D., Ph.D., who is currently a professor of internal medicine and biomedical engineering at the University of Michigan.
The article outlined miRNA profiling workflows for different sample types, emphasized the importance of quality control, and summarized the main miRNA quantitation techniques. The workflows encompassed:
- Purification techniques (such as gel electrophoresis, immunoprecipitation, and laser capture microdissection).
- Quality control checks (such as measuring spike-in oligo concentrations or housekeeping RNA expression,
spectrophotometric analysis, or automated capillary electrophoresis). - MicroRNA measurement techniques (such as quantitative reverse transcription PCR-based methods, hybridization-based methods, and RNA sequencing).
Tool Provider Commentary
To get a sense of how tool providers are helping researchers obtain reproducible results for miRNA quantitation, GEN’s editor in chief, John Sterling, posed a few questions to Douglas Horejsh, Ph.D., a senior research scientist at Promega, and Stephen Angeloni, Ph.D., a senior field applications scientist at Luminex.
GEN: What types of tools and reagents are critical for miRNA research?
Dr. Horejsh: Effective, safe, and reproducible miRNA purification tools are critical in the process. Obviously, it is very difficult to perform biomarker discovery if there is purification bias for particular miRNAs or inefficient dissociation of the miRNA from protein complexes. Promega has focused recent new product development efforts to purify total RNA including miRNA from tissues, plasma, serum, and exosomes. Many of these workflows are also automated, allowing for scaling of throughput to suit the needs of research laboratories.
Dr. Angeloni: For in situ hybridization detection of miRNAs, proper sample preparation and probes with good signal-to-noise ratios and specificity are critical. Several companies offer a variety of locked nucleic acid (LNA) probes for in situ detection of miRNAs along with protocols and reagents for sample processing. For PCR and sequencing technologies, the first limiting factor is the isolation of sufficient quantities of high-quality RNA from different samples. For some of these molecular technologies, the next critical factor is to have efficient amplification linkers or good stem-loop primers to accurately convert the miRNA into a cDNA that can be used for analysis.
GEN: What are some limitations of the tools and reagents currently being used in miRNA research?

Dr. Horejsh: The current standard method for purification of exosomes is ultracentrifugation. This method is reliable, but it is also tedious and time consuming. It is also not amenable to high throughput. Reagents that can be used to precipitate exosomes without ultracentrifugation would provide faster and automatable exosome purification. However, there appear to be differences in miRNA expression based on the exosome purification method. Additionally, most miRNA purification workflows currently on the market are completely manual. It is very difficult to scale this type of workflow to large biomarker discovery or research projects.
Dr. Angeloni: As more miRNA markers are associated with different diseases and become important in diagnostics, the need to detect more miRNA markers in small samples is increasing. For several PCR-based technologies, it is not easy to distinguish miRNAs that may differ by a single nucleotide. In addition, the quantity of high-quality RNA that can be isolated from various body fluids (urine, sera, other body fluids) can be too low to identify miRNAs at low concentrations accurately. When vital miRNA markers can vary by a single nucleotide, these factors can be a serious limitation for some diagnostic applications.
GEN: How are miRNA research tools and reagents being improved?
Dr. Horejsh: Promega will soon launch a Maxwell® purification kit for miRNA from plasma, serum, and exosomes. The Maxwell RSC is a small, automated instrument for 1–16 samples that performs many of the steps in the purification. The instrument purifies miRNA from a preprocessed sample without the need to perform manual binding, washing, and elution steps. The chemistry is an extension of our Maxwell RSC miRNA Tissue Kit that has changed the way that our collaborators are able to study miRNA expression. We are also looking at the possibility of moving this workflow to large automation.
Dr. Angeloni: Because of the diversity of miRNA expression patterns that can be associated with several cancerous and noncancerous diseases, the ability of technologies to detect different patterns of miRNA expression in samples of limited size is becoming more important. In addition, as with other diseases where expression patterns of markers may vary between individuals, the ability to analyze expression pattern differences between different individuals, for research and diagnostics purposes, requires the use of new assay technologies and bioinformatics analysis tools. In both areas, multiplexing assay platforms and new data analysis tools are providing solutions to these challenges.
GEN: MicroRNAs recently have been employed diagnostically using liquid biopsies. Are there any exciting other new applications for miRNAs on the horizon?
Dr. Horejsh: I believe that we are still early in our understanding of miRNA biology. Much as we’ve seen growing interest and utility of circulating cell-free DNA [cfDNA] in oncology research, miRNA will continue to grow in importance. I look forward to better understanding how exosomes, proteins, circulating cfDNA, and miRNA can be used to more universally study cancer early in disease course to tip the balance toward remission or cure over progression.

Dr. Angeloni: With changes in miRNA expression or sequence variations contributing to the development of different diseases, this opens the door for new therapeutic targets. As a result, the use of miRNAs as therapeutic agents has been tested with some success. Another exciting approach is the development of miRNA-targeting drugs. Several companies are developing drugs targeting miRNAs for cardiovascular diseases, some infectious diseases, metabolic diseases, cancer, and some other noncancerous diseases. Because miRNAs are involved with a broad range of human diseases, the development of new effective miRNA targeted treatment strategies has a bright future.
Circulating MicroRNAs as Biomarkers
A challenging but potentially rewarding application of miRNA profiling technology is to identify circulating biomarkers. Such biomarkers could be sought in patient samples obtained by noninvasive means, permitting repeat sampling and disease and treatment monitoring. As Dr. Tewari and colleagues noted in their review, however, “there is increasing concern around the need for rigorous control of preanalytic and analytic variables when considering potential circulating miRNA markers.”
This concern was mentioned in the review back in 2012, and it still pertains. To address this concern, Dr. Tewari recently led an effort to promote the reproducibility of liquid biopsy studies that quantitate miRNA by means of RNA sequencing.
As described in Nature Biotechnology, Dr. Tewari’s team prepared samples identically and sent them across the country for each of nine labs to analyze. Each lab used multiple testing protocols to sequence four different samples, including a plasma sample and three synthetic RNA samples. Altogether, the labs tested nine different sequencing protocols, including four commercially available kits and five protocols developed by the labs.
“Both protocol- and sequence-specific biases were identified, including biases that reduced the ability of small RNA-seq to accurately measure adenosine-to-inosine editing in miRNAs,” the article’s authors wrote. “We found that these biases were mitigated by library preparation methods that incorporate adapters with degenerate bases.”
“We realized,” said lead study author Maria D. Giraldez, M.D., Ph.D., a postdoctoral research fellow in Dr. Tewari’s lab, “that not only different methods produce different results, but also any small change within a given protocol can introduce an important degree of variation. To compare results across labs, it is key to use a common and highly standardized protocol.”
According to the study, different sequencing methods produced different and often inaccurate abundance estimates for individual markers. More encouragingly, the study also indicated that these estimates could be improved by using the methods developed by the consortium labs. Finally, as a University of Michigan press release noted, when RNA sequencing was used to compare the relative amounts of individual miRNAs between different samples, all the methods produced accurate and reproducible estimates.