April 1, 2014 (Vol. 34, No. 7)
Barrett Bready, M.D. Nabsys
John Thompson, Ph.D.
“One-Size-Fits-All” Sequencer Might Not Be in the Cards, but Customizable Solutions Could Be
With the advent of both Sanger and Maxam and Gilbert DNA sequencing technologies in the late 1970s, just the mere ability to sequence individual DNAs of interest was a great leap forward. This excitement led researchers to improve the initial technologies by focusing on reducing error rates, increasing read lengths, and making protocols robust enough for automation. Decades of improvements made sequencing much cheaper and powerful enough to tackle the human genome.
However, despite the vast improvements, sequencing remained focused on individual clones and DNAs. The next generation of sequencing changed that paradigm so that individual DNAs were no longer sequenced in a dedicated manner but, rather, sequencing targets were prepared in a massively parallel fashion and then sorted out after the sequencing was complete. This massively parallel approach resulted in instantaneous leaps in sequencing throughput and reduced costs.
Like the first-generation sequencing that preceded it, next-gen sequencing (NGS) has improved dramatically since its inception. Throughout the process of NGS optimization, the various commercial technologies offered different features and tradeoffs that users either knowingly, or in some cases unknowingly, chose among. The big three features that researchers found most compelling were cost per gigabase (Gb), read length, and error rate. One could never get the best of all worlds so users picked the combination that best suited their needs. When in doubt, users almost always defaulted to the cheapest sequencing method available at the time.
Many other features and issues were part of the discussion about the relative merits of different instruments. These included initial capital cost, ease of sample preparation, sequencing bias, amplification/single molecule, time to result, data handling/storage, and ability to detect base modifications. Ultimately, these factors were rarely a major factor in decision-making. The costs and throughput were changing so rapidly that laboratories and genome centers struggled to keep up with the ever-changing array of new and improved instruments.
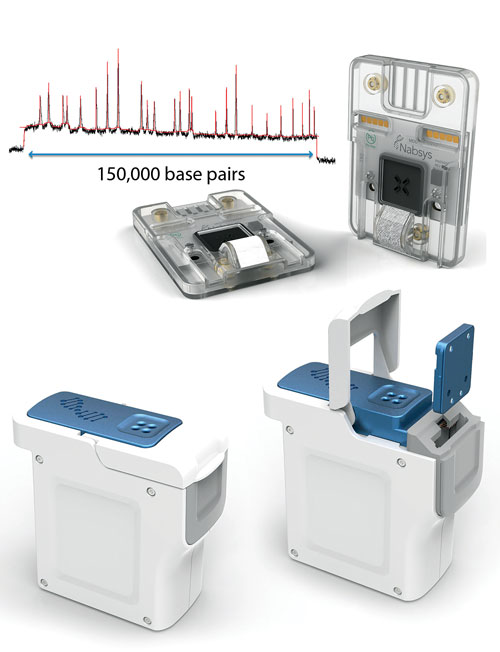
Single-molecule map of DNA from Nabsys semiconductor-based tools.
Factors beyond Price
However, the sequencing price freefall has stopped, or at least slowed, so that investigators have begun looking more closely at factors beyond cost; new instruments have moved in to take advantage of the needs of particular segments of the sequencing world.
For example, time to result can be critical in clinical sequencing, so taking a week just to carry out sequencing reactions and then having to analyze huge boluses of data is not an attractive option. Desktop sequencers that can turn around a sample in a day or so—but at a higher cost per Gb—have garnered a devoted following. Further advances in time-to-result and in decreasing instrument size will continue to be valued by those needing answers quickly
Indeed, the ultimate in reduced size has been actively discussed with excitement apparent for the development of portable, field-deployable instruments for sequencing samples in real-time or at remote/primitive locations.
In addition to smaller, more robust sequencing instruments, another critical need for next-gen sequencing is longer read lengths. De novo genome assemblies are still impossible with all but the smallest genomes due to repeat sequences that are longer than the read lengths. While de novo assemblies are not required in all projects, variation in copy number or genome structure can have significant impacts on data interpretation.
Multiple methods for generating real or virtual long reads have been described but none is currently available that achieves long enough reads at low enough cost to be satisfactory. Improvements in read length, either by direct sequencing or mapping very long molecules, continue to drive the development of new instruments, particularly those based on nanodetectors.
The high level of interest in the $1,000 genome and debate over the price point at which most people would want to get “their” genome sequenced obscures the fact that, in fact, no individual has a single genome that remains static over time. A low rate of mosaicism is expected across tissues in all individuals, and this variability can be very high in some cell types and will change continuously in tumors. Even more diverse and changeable is the complex set of genomes from microbes that inhabit our body surface and fluids. Thus, genomic data, though providing a solid foundation of understanding genetic effects, is only that—a foundation on which to place other genomic data.

Barrett Bready, M.D.
Viewing the Transcriptome
One particular type of genomic data that can provide a more dynamic view of an individual’s health and environmental status is an accurate view of the transcriptome in relevant tissues. Ideally, one would generate a complete picture of the transcriptome in all cells but this is not feasible for many reasons. Even if one can obtain the tissue of interest, analysis of all full-length RNAs cannot be carried out.
High-throughput methods can generate tags for all RNAs but may miss the variety of splice variants that occur in many species. Long-read technologies can generate a full-length read but current methods do not provide the read depth needed to understand the dynamics of poorly expressed genes.
Additionally, while long reads are critical for assembling genomes and generating accurate representations of long RNAs, many RNAs are too short to be effectively sequenced using standard protocols. Amplifying or ligating short RNAs can lead to highly biased results.
Thus, all RNA Seq methods in current use are a set of compromises in which one sacrifices information about the full range of the transcriptome being sequenced. More versatile sequencing methods may help extract a broader range of transcriptomic information.
In addition to simple sequence information, both DNA and RNA can also be modified and those methylations and more complex modifications can be functionally important. When DNA or RNA is amplified for sequencing, this information is frequently lost. Single molecule methods in which the original nucleic acid is sequenced directly can retain this potentially important information.
Improvements in technologies to gather this information would help us to better understand the importance of epigenetics, an area in which we have only begun to scratch the surface.
While the cost issue with sequencing has, to a first approximation, been solved, nothing is ever cheap enough. There are still many projects limited by how much sequencing reagents cost so having methods that are cheaper and easier will continue to drive new instrument development. Of particular interest now and in the near future are the many nanodetector technologies where sample preparation can be simplified and reagent and instrument costs reduced by using electronic detection and single molecule methods.
No sequencing instrument will provide the diverse kinds of data needed by all researchers. The future of sequencing will not be a single type of monolithic sequencer sold to large genome centers but, rather, will include a much more distributed sequence capacity tuned to the scientific or clinical needs of local investigators.

John Thompson, Ph.D.
Barrett Bready, M.D. ([email protected]), is president, CEO,and co-founder of Nabsys. John Thompson, Ph.D., is the company’s director of assay development.