April 1, 2006 (Vol. 26, No. 7)
Pure Production and Stable Isotopic Labeling of Desired Proteins with the SPP System
Elucidation of protein structure and function maintains an important role in post-genomic sequencing and analysis studies. An efficient protein production system is critical for obtaining large amounts of correctly folded recombinant protein for study. Escherichia coli, or E. coli, expression systems are used extensively for production of recombinant proteins and have two major advantages over other expression systems—ease of use and low cost. Some recombinant proteins, however, do not fold correctly during expression in E. coli, resulting in deposits of inactive insoluble proteins, or inclusion bodies.
In addition to target protein solubility, yield, purity, or signal-to-noise ratio, is also of importance. Using the conventional T7 promoter method in E. coli, approximately 30% of the soluble protein can be expected as the desired product.
In collaboration with Masayori Inouye at the University of Medicine and Dentistry of New Jersey, Takara Bio (www.takarabio.com) developed a novel system for generating large amounts of soluble and pure protein. The system uses cold shock technology and an endoribonuclease, mazF. The SPP System utilizes a modified version of existing cold shock expression vectors and is ideal for high-purity production (90%) and stable isotopic labeling of the desired protein.
Although cold shock expression vector systems allow for stable isotopic labeling in a desired protein-specific manner, the SPP System can suppress expression of proteins other than the desired protein more effectively and provides higher purity and labeling efficiency for a wider variety of target proteins. This suggests possible applications for protein structural analysis by NMR or other technologies.
Cold Shock Technology
The pCold DNA Vectors are a series of unique protein expression vectors, providing increased in vivo protein yield, purity, and solubility of expressed recombinant proteins using cold shock technology. More specifically, the cspA (cold shock protein A) promoter and related elements have been incorporated into these vectors to up regulate target protein production at lowered incubation temperatures, ranging between 37C-15C. The temperature drop also suppresses expression of other cellular proteins, represses protease activity, and temporarily halts overall cell growth.(1)
This process allows for high-yield, high-purity (up to 60% of intracellular protein; 50-400 mg protein/L culture) expression of target proteins and increased solubility as compared with conventional E. coli expression systems. Additionally, these vectors work well for synthesis of proteins to be labeled isotopically.
The method for protein production using pCold vectors is not much different than conventional methods (e.g. T7 vectors). In the example described here, a target gene is cloned into the multiple cloning site of the pCold expression vector. The host E. coli strain is then transformed with the expression plasmid and an ampr transformant selected. After inoculation with ampicillin, the culture is shaken at 37C until an OD600 of 0.4-0.5 is reached. Once this desired OD600 is obtained, the culture is refrigerated for 30 minutes at 15C without shaking, initiating the cold shock. To induce protein expression, IPTG is added and the culture is left shaking at 15C for 24 hours.
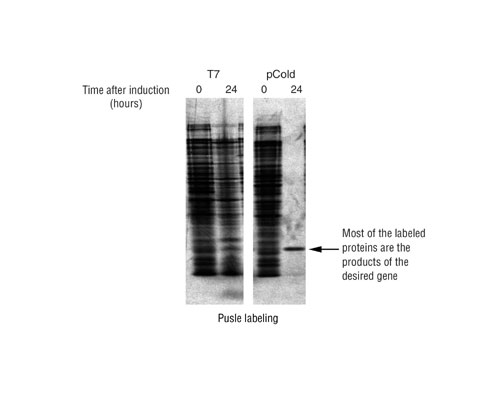
The cells are collected and target protein expression is confirmed with SDS-PAGE. Figure 1 shows results obtained when pCold vectors are used under these conditions.
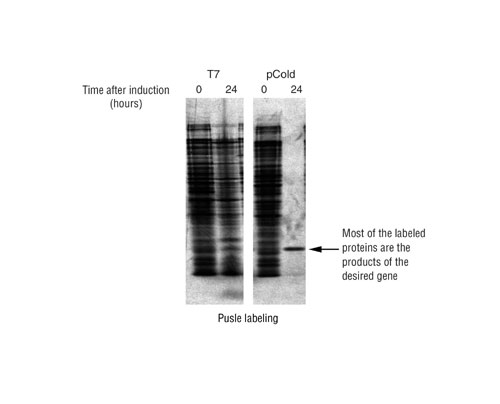
Fig.1: A comparative study of pulse labeling of human gene D protein using the cold shock expression system and the T7 expression system was performed.
mazF Endoribonuclease
Suicidal genes (toxins) are suspected to cause permanent damage to cells, leading to programmed cell death, or stasis. E. coli contains at least five such toxin-classified genes—chpBk, mazF (chpAK), rel E, yafQ, and yoeb. These five genes are co-expressed with their associated antitoxin, and under normal cellular growth conditions, this co-expression prohibits the toxin effect.
Research has shown that one of these toxins, mazF, possesses mRNA endoribonuclease activity and is very similar in enzymatic function to RNAse A. mazF specifically cleaves single-stranded cellular RNA at the 5´end of ACA sequences. The enzyme does not cleave double-stranded RNA nor does it cleave single- or double-stranded DNA. Cellular rRNAs and tRNAs are able to circumvent mazF cleavage activity because of rRNA affiliation with ribosomal proteins and the considerable amount of secondary structure of tRNAs.
The endoribonulease cleavage occurs at the same site within the ACA sequence whether the RNA is in vivo or in vitro. Upon cleavage of the mRNA the enzyme leaves a 2´-, 3´-cyclic phosphate and a free 5´OH group. The enzyme function is not limited to substrate size, indicated by its ability to cleave a synthetic RNA, as small as seven bases in length. mazF, with regard to all of its functional characteristics, behaves as an RNA restriction enzyme.(2)
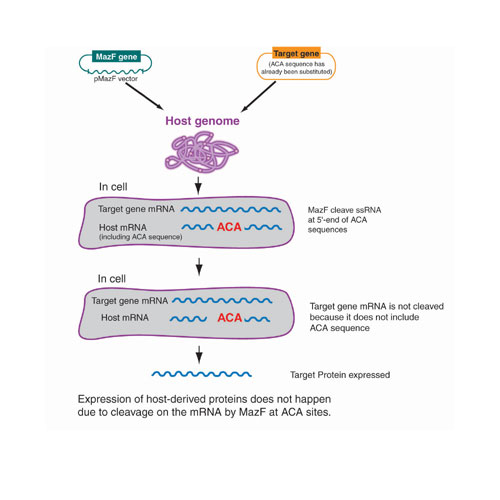
Fig.2: SPP System(tm) (Single Protein Production System
Single Protein Production (SPP)
The SPP System consists of two plasmids that are co-transformed for expression. One vector carries the E. coli toxin protein, mazF, and the other is a modified pCold vector that does not contain any ACA sequences. As previously stated, the mazF protein was determined to be a sequence-specific endoribonuclease that cleaves single-stranded RNAs at ACA sequences. In order to effectively use this system, the transcript of interest should not contain any ACA sequences; i.e., should be ACA-less.
The ACA-less gene of interest can be constructed by chemical synthesis or site-directed mutagenesis technology. In designing the ACA-less gene, every ACA sequence within the gene of interest must be modified to make its amino acid sequence and restriction sites for cloning in frame. This ACA-less gene is then cloned into pCold (SP-4) vectors, which are ACA-less pCold vectors.
An E. coli host is then co-transformed with this expression plasmid and the pMazF expression plasmid. Expression of mazF is induced, cleaving all other cellular mRNA derived from the host proteins or others at ACA sequences but leaving the target mRNA intact. The result is highly pure (up to 90% of cellular protein) target protein expression with almost no cellular protein background. Despite the stress of undergoing a drop in growth temperature, target protein synthesis can be detected for up to four days.(3,4) Figure 2 illustrates the design of the SPP System.
To demonstrate the viability of the system for structural analysis studies, an experiment using the envZB gene was performed. A control expression plasmid was prepared by inserting an ACA-less ORF of E. coli-derived envZB protein into pCold I (SP-4) DNA. The estimated molecular weight of the expressed protein is 19.6 kDa. E. coli BL21 cells were co-transformed with pCold I (SP-4) vector and pMazF in 3 mL of M9-glucose medium with 50 µg/mL ampicillin and 34 µg/mL chloramphenicol at 37C.
Once the OD600 reached 0.5, the culture was placed at 15C for 45 minutes in order to facilitate cold shock expression. mazF and envZB induction was initiated with the addition of 1 mM IPTG. After 24 hours of induction, 250 µL of the culture was added to 19 amino acids (50 µg/mL each, minus methionine) and 15µCi [35S]-methionine. At the end of a 15-minute isotopic labeling at cold shock temperatures, nonradioactive methionine was added and incubated for an additional five minutes.
Labeled cells were washed in a Tris-Cl, NaCl buffer and then centrifuged to remove the buffer. The pellets were resuspended in SDS-PAGE loading buffer and resolved on 15% SDS-polyacrylamide gel. After drying, the gel was exposed to X-ray film overnight. Results are shown in Figure 3.
The pulse labeling results demonstrate that using the SPP System, a union of pCold vectors and mazF, up to 90% of the newly synthesized protein expressed is the protein of interest. mazF degraded host proteins mRNAs, resulting in low cellular protein background and enhancement of the ACA-less target protein.
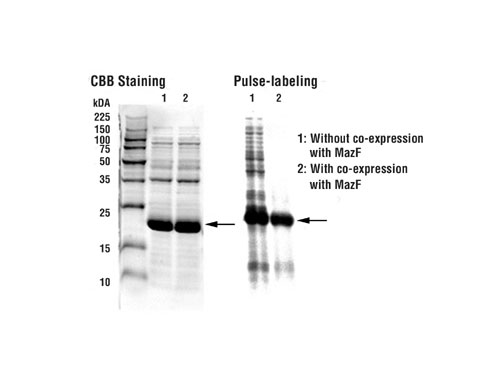
Fig.3: The pulse labeling results demonstrated a reductin in background host protein levels with mazF coexpression.
Conclusion
The SPP System offers a novel approach to protein production. Research demonstrates that the union of cold shock technology and the endoribonuclease mazF provides several advantages over other bacterial expression systems. The system provides nearly exclusive isotopic labeling of a target protein for NMR and X-ray crystallography structural analysis. NMR structural analysis can be performed in vitro and in a protein’s natural environment. In addition, it allows for structural studies to be conducted on proteins that are toxic to other systems. Also, the system offers a cost-effective alternative to other more labor intensive and expensive in vitro expression systems.(3,4)