Tracy Vence GEN
Using metabolic phenotyping approaches, researchers are deconstructing gene-environment interactions and their contributions to disease risk at the personal and population scales.
Increasingly it is recognized that when it comes to common diseases, the genome can only help solve part of the puzzle. With conditions like hypertension, for example, there may be several genetic variants that each have some small effect on an observed phenotype—elevated blood pressure. But even when taken together, genetic variation alone cannot fully explain why some people develop hypertension while others do not.
Long before scientists sought out the genes behind hypertension, they recognized how environmental triggers can raise blood pressure. They identified physiological and biochemical markers indicating as much.
Now, considering both genetic and environmental influences, researchers are bridging the gap between nature and nurture, learning more than ever before about how gene-environment interactions cause—and may be predictive of—disease.
“Through life, there’s a series of conditional gene-environment interactions that shape your future and your likelihood of getting a disease, and also change the way you respond to therapies when you do have a disease,” says Imperial College London’s Jeremy Nicholson, Ph.D., head of the department of surgery and cancer. “I think there’s a general realization that we have to find new metrics for the environmental interaction and that we do have to figure out how that maps onto human genetic variation.”
Dr. Nicholson and his colleagues are putting metabolic phenotyping techniques to the test on two very different scales—at the level of individual patients with an eye toward tailored treatments, and broader efforts, encompassing entire populations in epidemiological studies.
Personal Phenomics
Perhaps at no other time is it more critical to be able to distinguish healthy tissue from cancer than when a patient is on the operating table. As malignancies are not always discrete, it can be tough for the surgeon to tell where and how much to cut.
It’s typically rare to find a mass spectrometer in the OR, but if the results of a study published in Science Translational Medicine today hold up in other hospitals, it’s not hard to imagine the trusty machine could become commonplace there.
Dr. Nicholson and his colleagues at Imperial have created what they call an “intelligent” surgical knife, or iKnife, which couples rapid evaporative ionization mass spectrometry (REIMS) to electrosurgical dissection, and enables tissue identification in real time. The iKnife works by analyzing the surgical smoke produced by cauterization via REIMS, generating disease state-specific metabolic profiles.
Writing in Science Translational Medicine, the researchers report having applied this approach to analyze in the lab a variety of tissue samples from 302 patients. They also used the mass spec-coupled iKnife to analyze 81 surgical resections in theater—that is, performed in the OR. The researchers compared their metabolomic findings against the results of traditional, post-operative histopathological analyses, confirming the accuracy of their technique. Of the 81 resections performed, 100% of the intraoperative REIMS results matched the classic histology-based readouts. “These data provide compelling evidence that the REIMS-iKnife approach can be translated into routine clinical use in a wide range of oncosurgical procedures,” the researchers conclude.
According to Dr. Nicholson, the technique has been successfully applied to more than just cancers.
“We connect the exhaust pipe from the knife to a mass spectrometer and we can do tissue diagnostics in less than half a second, displaying the data that comes out of the iKnife mass spectrometer to the surgeon in real time,” Dr. Nicholson tells GEN. “It can give you information about tumor margin, to cut or not to cut, tissue viability, ulcerative colitis versus Crohn’s disease, cancer margins, different sorts of lymph nodes—it gives you an enormous range of background biochemical information in a surgical decision timeframe.”
The researchers have even used the iKnife to distinguish horsemeat from beef.
“It’s an absolute total revolution,” he adds, “not only in terms of the technology, but also the information delivered based on metabolism.”
Dr. Nicholson says future plans include to development and optimization of an intelligent endoscope, which could help physicians make treatment decisions related to lung biochemistry, for example. “Some of the technology we have developed for surgery actually can be redeployed in a different way for looking at other sorts of analytical problems, in fact any sort of complex mixture analysis in this way,” he notes.
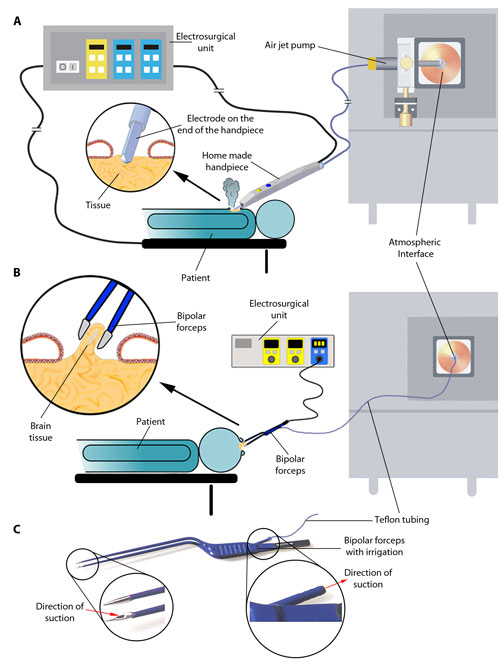
Schema of REIMS instrumentation and data collection. (A and B) Surgical ion source and ion transfer setups for REIMS experiments using monopolar electrosurgery (A) or bipolar electrosurgery (B). (C) Schematic of aerosol aspiration using commercially available bipolar forceps. [Science Translational Medicine/AAAS]
Population Scale
As the 2012 Olympic Games were winding down, so too was the need for a colossal drug-testing facility. London’s Olympic drug-testing operation housed swaths of analytical chemistry equipment, which, initially intended to screen althletes for performance-enhancing compounds among other things, found a new home in a large-scale phenomics facility once the sporting events were over.
With £10 million (approximately $15 million) in funding from the U.K.’s Medical Research Council and National Institute of Health Research to support the repurposing and initial use of these machines, plus additional equipment supplied by Bruker Biospin and Waters, the MRC-NIHR Phenome Center was born.
“Just like large-scale genomics requires lots and lots of next-generation sequencers in a row, large-scale metabolic phenotyping requires lots and lots of technology in a row so they can process vast numbers of samples and so you can start tackling those problems of population physiology and biochemistry, which haven’t really been possible until recent years,” says Dr. Nicholson, who heads the now month-old facility.
“What we’re interested in doing is…metabolic phenotyping from samples people in the general population and trying to link those variables into, say, diastolic blood pressure, so you can understand which factors in metabolism might be from exogenous exposures might be related to that generalized disease risk,” he explains. “If you’ve got tens of thousands of samples, you need a lot of analytical chemistry to be able to do this.”
Pilot projects currently being run at the center run the gamut from discerning type 2 diabetes risk in Indian-Asian populations, to understanding how metabolic phenotypes associated with hypoxia could inform improved anesthetic practices. “The information generated here is to inform long-term policy about the health of populations,” Dr. Nicholson says.
Another goal of the center is to train future scientists and physicians in how to harness the phenome at the population scale in an effort to fill the gaps left, for example, by genome-wide association studies (GWAS).
“In almost all these cases, the samples we get [come] with genomic information. There will be either GWAS-type information…maybe exome sequencing data as well. By doing that, of course, we get the true genetic underpinning and variation,” he says. “We also have the metabolic outcome and phenotype, which links it [genomic information] to environment. By linking those together, you can do deep gene-environment deconvolution for risk factors, for the first time.”