September 1, 2017 (Vol. 37, No. 15)
Kate Marusina Ph.D.
Using CRISPR to Produce Better Disease Modelling
Tricks of the genome-writing trade were freely exchanged at the first annual Genome Writers Guild conference entitled, “Realizing the Future: Genome Engineering,” which was held at the University of Minnesota in July 2017.
Besides suggesting ways to release genome-writing creativity, speakers at the meeting discussed technical advances, potential biotechnology applications, and prospects for clinical translation. The meeting even included TED-style talks and a science fiction writer’s views on genome engineering’s utopian (and dystopian) possibilities.
The bulk of the meeting, however, focused on more immediate technical issues. Most sessions focused on the latest genome-engineering tools and their diverse applications, from dissecting the cell’s molecular mechanisms, to harnessing these mechanisms for whatever purposes that are deemed worthy.
Genome editing is giving us the power to change the world, a power that needs to be wielded responsibly. For example, therapeutic and reproductive genome editing have tremendous potential to alleviate illness, but we need to ask if this potential be realized safely, without unintended consequences.
High-Fidelity CRISPR Toolbox
“CRISPR gene-editing technology is taking biology by storm,” points Mark Behlke, M.D., Ph.D, chief scientific officer, Integrated DNA Technologies (IDT). “The CRISPR toolbox consists of three primary components: a guide RNA, which targets a specific locus in the gene; a Cas9 nuclease, which creates a DNA break at the locus; and a DNA repair template, which enables gene replacements and insertions. IDT holistically optimized the entire toolbox, resulting in a highly efficient, error-resistant system.”
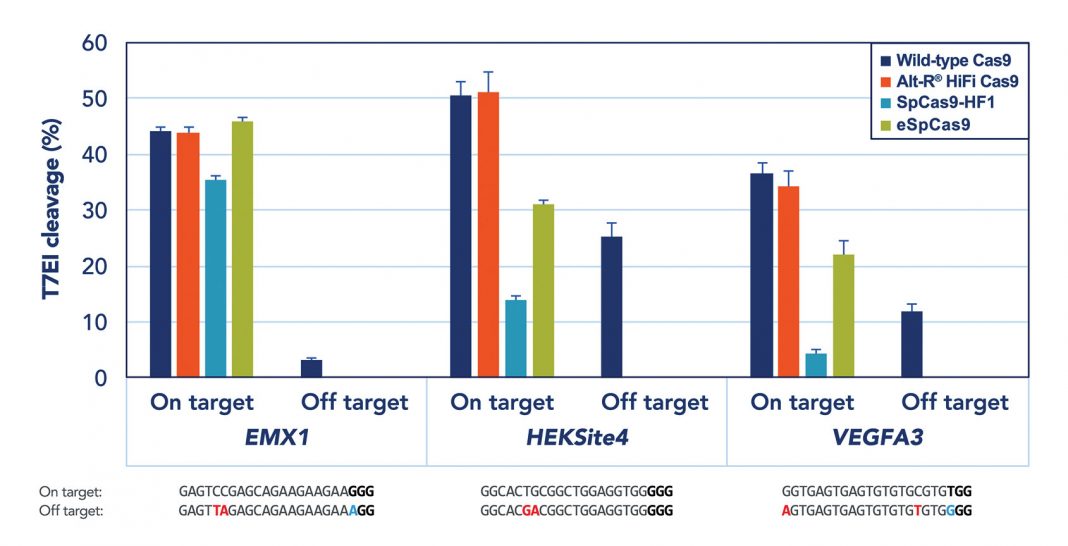
While there are numerous approaches to implementation of CRISPR/Cas9 gene editing, many of them experience a relatively high rate of off-target effects, limiting the system’s applicability. Dr. Behlke explains that if Cas9 is expressed from DNA or even messenger RNA, the enzyme persists in a cell for a long time, increasing chances of cleavage at nonspecific sites.
IDT and others have demonstrated that a ribonucleoprotein (RNP) complex consisting of Cas9 and guide RNA could be delivered at high concentration, but degrades rather quickly. “The RNP approach limits the amount of catalytic components and gives [a] “fast-on/fast-off” character to the genome-editing machinery,” says Dr. Behlke. Moreover, IDT created chemically modified derivatives of the two components of the guide RNA to reduce the risk of triggering the innate immune system.
This highly optimized RNP complex could be delivered by any standard transfection technique. Microinjection of Alt-R® CRISPR/Cas9 RNP system is particularly valuable for generation of knockout and transgenic mice. Electroporation of primary T cells with Alt-R RNP complex targeted to alpha chain of the T-cell receptor routinely results in over 80% knockout of surface TCR complex, without detectable toxicity (Figure).
“Our next significant improvement is a novel Cas9 mutant,” continues Dr. Behlke. Using directed evolution, scientists developed a high-fidelity Cas9 mutant that fully retains on-target activity with significantly reduced off-target activity. The Alt-R RNP complex—combining chemically modified synthetic guide RNA and the high-fidelity Cas9 mutant—brings CRISPR technology even closer to clinical applications.
Using IDT reagents, a team of Stanford University scientists led by Matthew Porteus, M.D., Ph.D., demonstrated robust results in correcting sickle-cell mutations in bone marrow stem cells, potentiating translation into clinical trials. Such developments, suggests Dr. Behlke, should justify an optimistic outlook: “Precision editing of [the] genome for human health is not such a distant reality.”
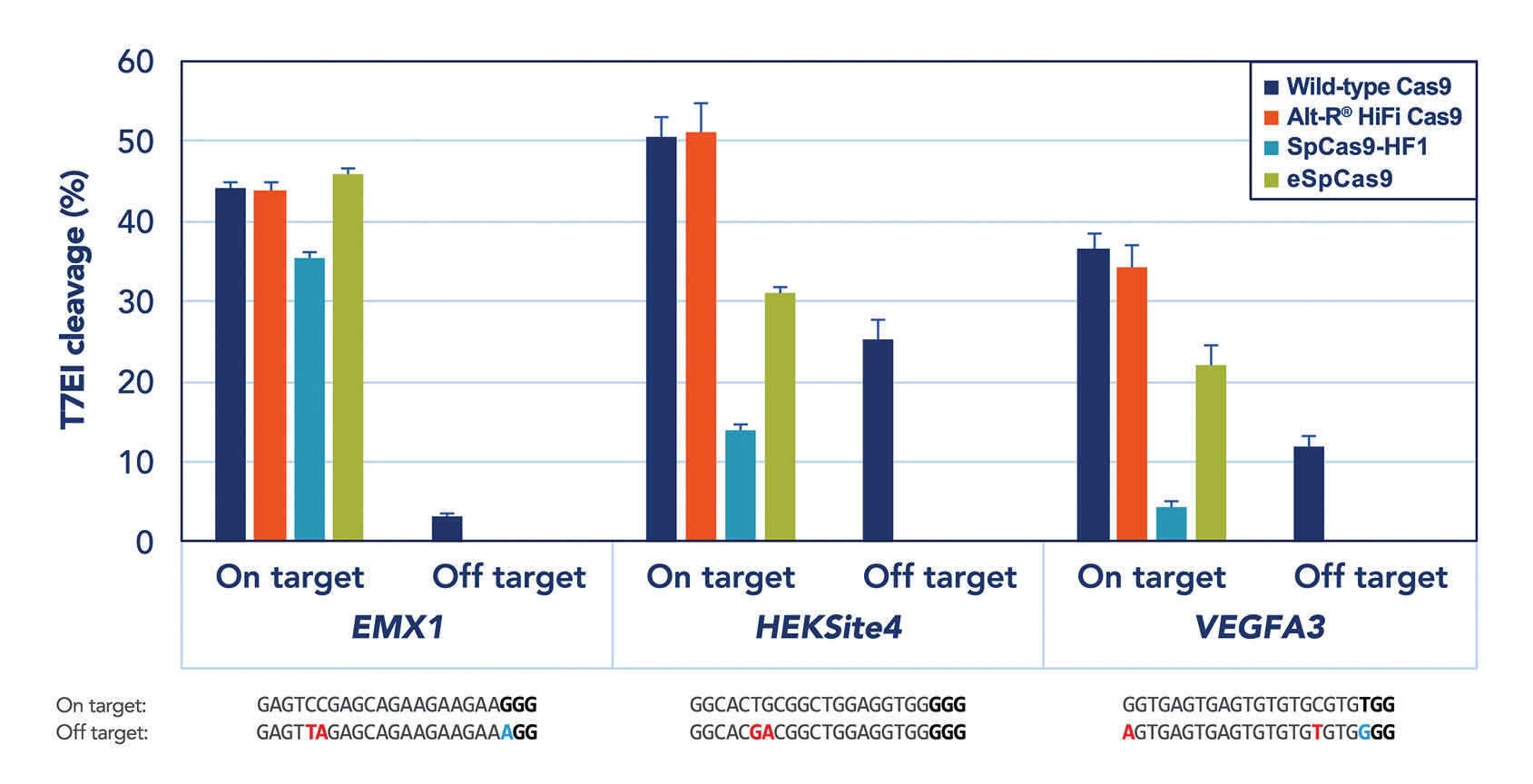
Figure. To address concerns about the off-target editing commonly observed in CRISPR/Cas9 applications, Integrated DNA Technologies has developed a high-fidelity, Streptococcus pyogenes Cas9 nuclease. When delivered as a ribonucleoprotein, this nuclease, called Alt-R® S.p. HiFi Cas9 Nuclease, can provide superior on-target performance, maintaining the high potency of wild-type Cas9 while reducing off-target editing.
Zebrafish Genome Explains Hearing Loss
The human sense that most commonly becomes impaired is hearing. In the United States alone, reductions or losses in the ability to detect sounds have been experienced by approximately 28 million people.
When hearing becomes impaired, the underlying cause is usually (more than 50% of the time) genetic. Various genetic causes of hearing impairment are of interest to Lisa A. Schimmenti, M.D., a medical geneticist at the Mayo Clinic Rochester.
For example, she investigates genetic variations that affect a protein called gap junction beta-2 (GJB2). These variations account for more than 50% of hearing loss cases worldwide that are nonsyndromic (i.e., not associated with other symptoms), and for about 25% of such cases among children in North America. Families carrying GJB2 variants demonstrate considerable spread in phenotype from completely deaf to mild impairment, suggesting other contributing factors.
To model human hearing, Dr. Schimmenti’s laboratory relies on the zebrafish. “The zebrafish is a practical model organism for creating genetic mutations on the basis of human variants,” says Alaa Koleilat, a Ph.D. student at the Mayo Clinic Rochester. “The zebrafish genome contains [a] high percentage of related orthologue human genes.”
Continues Koleilat, “Additionally, zebrafish have a series of mechanosensory receptors similar to human hair cells, easily accessible in the fish lateral line. Most importantly, deaf zebrafish display a characteristic circular swimming behavior, making it straightforward to distinguish [them] from wild type.”
Dr. Schimmenti’s team hopes to create model organisms for discovery of pharmacological treatments for hearing loss. Using a gene-editing technology based on transcription activator-like nucleases (TALENs), the team has modeled the most common human variant in GJB2, c.35delG.
“Unlike the CRISPR system, the TALEN system is unrestricted in terms of the sequence types it can modify,” notes Koleilat. “A TALEN is guided by its own DNA-binding domain, which can be precisely engineered for a target DNA sequence. TALENs can be developed in less than a week.”
The team created a mutant line with an 8-bp out-of-frame deletion that results in an early stop codon in the zebrafish orthologue of GJB2, cx30.3. Dr. Schimmenti explains that in situ detection of the protein is challenging because the protein itself is small.
“So far, we observed that homozygous recessive embryos have decreased expression of cx30.3 in the ear and no expression behind the heart,” reports Dr. Schimmenti. “And yet these embryos display normal hair cell structure and also have normal mechanotransduction channel activity.” A subset of homozygous embryos demonstrates circling behavior linked to deafness.
The team continues to perform functional validation studies in knockout model of cx30.3. Going forward, the team hopes to extend its genetic-engineering methodology to model other genetic variants associated with hearing and vision loss in humans.
Novel Vector Suite for Precise Integration
Gene knockout and knockin are critical techniques in functional gene analysis, and they may become standard approaches in gene therapy. These techniques have already demonstrated greater utility, given their incorporation of custom CRISPR/Cas9 nuclease systems.
CRISPR/Cas9-based knockout and knockin techniques are being developed by Drena Dobbs, Ph.D., Jeffrey Essner, Ph.D., and Maura McGrail, Ph.D., Iowa State University scientists who specialize in bioinformatics, cellular biology, and genome dynamics, respectively. These scientists are collaborating not just with each other, but also with Stephen C. Ekker, Ph.D., and Karl J. Clark, Ph.D.—Mayo Clinic researchers who apply genome engineering to zebrafish models. (These researchers also work on behalf of Lifengine Technologies, which provides gene-editing reagent kits.)
“We built on an in-frame integration method that was recently described in Scientific Reports, in a paper credited to Hisano et al., informs Jordan M. Welker, a graduate student at Iowa State University. “When this method is used, a reporter gene flanked by short regions of homology can be efficiently integrated at a DNA double-strand break using CRISPR technology.”
“The Essner, McGrail, and Dobbs laboratories, in collaboration with the Clark and Ekker laboratories, created specialized vector technology that enables precise knockins. The key is in the design of the short homology arms. Because the arms are 24 to 48 bases long, they ensure high integration efficiency. Altering donor homology lengths by as little as one base pair can double knockin efficiencies.”
The technology is called pPRISM, for PRecise Integration with Secondary Markers. It contains sites for cloning gene homology arms, options for several colors of fluorescent tags, or a stop codon. Homology arms are designed to match the sequences directly upstream and downstream of a CRISPR genomic DNA target site.
The Iowa State University/Mayo Clinic team also designed a “universal gRNA” that can efficiently liberate the knockin cassette from the vector and expose the regions of homology. pPRISM is optimized for the Golden Gate cloning method, where multiple DNA fragments can be directionally assembled into a vector backbone in a “one-pot” reaction. In vivo targeting with pPRISM and CRISPR/Cas9 directs precise knockin with little mosaicism in up to 80% of injected animals.
Welker uses the technology to study connexin 43.4, a membrane protein involved in intercellular communication and left/right development. “The pPRISM technology enabled knockin of specific connexin 43.4 alleles tagged with different fluorescent markers,” says Welker. “We can then follow each allele to ascertain its effect on heart laterality.”
The Essner and McGrail laboratories successfully targeted six loci in the zebrafish genome and will make this technology available to the community. “We expect our vectors, protocols, and web interface for homology design to streamline experimental design and broaden the use of CRISPR for homology-based gene editing,” adds Welker.
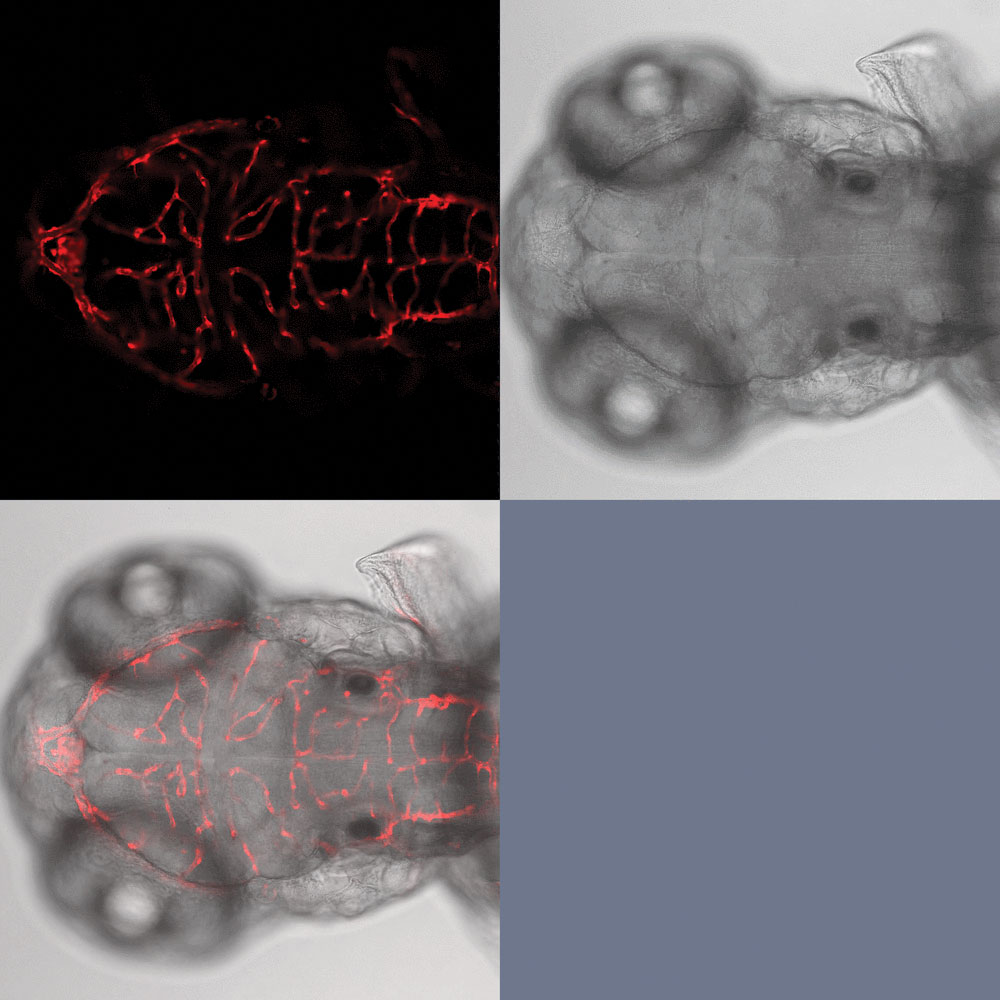
A team of Iowa State University scientists is dev- eloping specialized vector technologies that enable precise knockins. This confocal image, provided by the team’s Wesley Wierson, shows a zebrafish embryo for which a fluorescent reporter has been integrated (at the esama locus).
Natural Killer Cell Therapy in Action
Natural killer (NK) cells, cytotoxic lymphocytes of the innate immune system, identify target cells—virally infected cells and cancer cells—by recognizing complex patterns of membrane receptors. Once they have identified their targets, the NK cells live up to their name.
Multiple therapeutic approaches have attempted to harness NK cells. For example, clinical trials have been initiated to determine whether adoptive cellular transfer of allogeneic NK cells could be effective against solid tumors such as neuroblastoma, renal, colon, gastric, and ovarian cancers. In these trials, NK cells tend to disappoint. They may lack efficacy because they are negatively downregulated by the tumor microenvironment.
“If we are able to knockout these negative regulators, we can create an NK cell that is equipped to fight cancer,” says Emily Pomeroy, a graduate student in the laboratory of Branden Moriarity, Ph.D., a pediatric oncologist at the University of Minnesota. “Combined with easy expansion and well-developed adoptive transfer, such modified NK cells could become a single source of immunotherapy to treat a broad range of cancer types.”
The research team, led by Dr. Moriarity, used targeted genome editing to knockout genes in primary immune cells, including NKs, which are notoriously difficult to edit. (As part of this effort, Pomeroy helped to develop a transfection protocol. Specifically, she led work to define electroporation conditions that are capable of preserving cell viability while maintaining 95% transfection efficiency.) Five known downregulator genes were selected, and knockouts were fully characterized.
The team plans to put together a combination of edited NK cells to achieve maximum antitumor efficiency. “A significant advantage of NK cell therapy is that no matter how potent it is, the transplanted cells do not persist,” added Pomeroy. In comparison with antibody blockade of inhibitory receptors, another considerable advantage of CRISPR-mediated gene editing lies in its ability to knockout intracellular proteins.
Dr. Moriarity’s team has already created multiplex knockouts. Going forward, it hopes to expand on its work with CRISPR-edited NK cells. Soon, these cells may be evaluated in vivo, in studies using human cancer cell lines xenographed into mice. Ultimately, it is feasible that the team’s work could find its way to the clinic.
Examining FnCas9
The CRISPR/Cas9 system has greatly enhanced gene editing in eukaryotic organisms. However, there is a continued need to improve both cleavage activity and guide RNA design density and specificity.
In the exploration of CRISPR systems, MilliporeSigma scientists report that they have observed type II-B FnCas9 from Francisella novicida to possess a higher intrinsic specificity than SpCas9.
Motivated by its high specificity, the investigators set out to examine the robustness of FnCas9 in comparison with SpCas9. In contrast to SpCas9, FnCas9 activity fluctuated greatly along several genomic regions and failed to cleave a large number of the targets in chromosomal regions at which SpCas9 cleaved efficiently, notes Martha Rook, Ph.D., head of gene editing and novel modalities at MilliporeSigma, who adds that regional variability in endonuclease activity is not exclusive to FnCas9.
“We also observe type II-C CjCas9 and NcCas9 and type V FnCpf1 to display little or no cleavage at an array of genomic loci,” she points out. “Several studies suggest that chromatin structures can be a major barrier to RNA-guided endonuclease binding and cleavage in mammalian cells.”
After observing consistent activity on purified DNA substrates and testing variations in guide RNA length at cleavable and apparently uncleavable genomic sites, the team reasoned that target-site accessibility might be the limiting factor in editing efficiency in these cases.
“Our scientists hypothesized that the binding of an orthogonal, catalytically dead S. pyogenes Cas9 (SpdCas9) at proximal locations might alter local chromatin structures and render otherwise inaccessible target sites now accessible and subsequently cleavable by alternative nucleases such as those mentioned above,” continues Dr. Rook.
“In a recent publication [Chen, et al., “Targeted Activation of Diverse CRISPR-Cas Systems for Mammalian Genome Editing via Proximal CRISPR Targeting,” Nat. Commun. 8, 14958 (2017)], we show a restoration of alternative nuclease activity by co-targeting SpdCas9 to bind at proximal sites. Our scientists indicate that this proxy-CRISPR targeting strategy is applicable to a range of genomic loci and as a proof of concept, shows that the strategy may enable selective editing between identical genomic sites by judiciously selecting proximal dCas9-binding sites to differentiate the identical genomic locations.”
These findings provide a method to improve both genome-editing precision and efficiency using diverse CRISPR systems and a path to a better understanding of the impact of chromatin microenvironments on genome modification, states Dr. Rook.
Continue reading the complete set of Genome Engineering articles.
A Guild for Sharing Genomic Know-How
CRISPR Proving More User-Friendly
Center Positions Minnesota as North Star of Genome Engineering