April 15, 2017 (Vol. 37, No. 8)
DeeAnn Visk Ph.D. Founder and Principal Writer DeeAnn Visk Consulting
To Head Off Cancer at the Pass, Take Directions from Pathway Analysis
Almost 100 years ago, German biochemist Otto Heinrich Warburg noticed that cancer cells tend to have a high rate of glycolysis compared to normal tissues. This phenomenon, which occurs in cancer cells along with increased glucose uptake and fermentation of glucose to lactate, is known as the Warburg effect. Present-day explanations for the Warburg effect remain unsettled.
Interestingly, a 2016 article in Trends in Biochemical Sciences concluded that advances in cancer therapies that target metabolic pathways would aid in understanding the Warburg effect. This article will discuss progress in finding some of these metabolic drug targets.
A vast body of literature on the metabolic pathways and biochemical reactions has been generated from research done over many decades. Mining this literature is one method to deduce potential metabolic targets for new cancer therapies. Another approach is to metabolically screen cells before and after treatment, or to analyze cancer cell metabolism and compare it to the metabolism of normal cells.
“Cancer cells require lots of energy to grow and proliferate, and many pathways get co-opted into providing for these bioenergetic and biosynthetic needs,” says John Ryals, Ph.D., president and CEO of Metabolon. Advances in mass spectroscopy techniques and data analysis greatly improve the ease of obtaining metabolic data from cells. “We need only about 10–25 mg of a cell pellet to determine the metabolic profile of the cells,” continues Dr. Ryals.
In addition to studying cancer, Metabolon investigates many other metabolic diseases. Recently, company representatives contributed to a study demonstrating that metabolomics could be an effective tool in precision medicine for disease risk assessment and customized drug therapy in clinics. This study, which appeared in the Proceedings of the National Academy of Sciences, identified metabolic abnormalities consistent with early indications of diabetes, liver dysfunction, and disruption of gut microbiome homeostasis.
“On a certain level, all diseases are metabolic,” comments Dr. Ryals. “They are a physical manifestation of what is going on with a patient. Looking for functions of genes (for which the function was previously unknown), we collaborated with companies such as Human Longevity, bringing together genomic and metabolic data, to find the function of these genes.
“These initial studies have analyzed the genomes and metabolomes of 2,000 to 3,000 people, and from those large sets of data, we’ve been able to discover the function of about 200 to 300 genes. With this new knowledge, we look forward to a day when there is enough genomic and metabolomic information on much larger numbers of people to give us an even clearer picture of how this all fits together. Someday, we will be able to understand the function of all genes and where their metabolic outcomes lead.”
Opting for Strategic Interventions
Drug candidates that function by modulating metabolism do not have to be identified through complex screens of the metabolome. If the literature is reviewed with an eye toward finding reactions that are essential for metabolic processes, ideas can be generated for drugs that work via novel mechanisms of action to impinge on metabolic reactions.
The latter approach—reviewing pathway knowledge to develop mechanistic insights and identify strategic opportunities—was followed by Paul Bingham, Ph.D., vice president of research at Cornerstone Pharmaceuticals and professor of biochemistry and cell biology at Stony Brook University. Mechanistic insights helped Dr. Bingham and colleagues develop a first-in-class drug that attacks core mitochondrial metabolism in tumor cells.
“We were able to tap into the vast knowledge base of metabolism to observe where regulatory processes are reconfigured in tumor mitochondria, inviting cancer-specific targeting,” recalls Dr. Bingham. “These observations suggested that a lipoate analog can directly address lipoate-dependent, tumor-specific regulatory targets, resulting in efficient, selective tumor cell death.
A lipoate-containing enzyme, pyruvate dehydrogenase (PDH), is central to metabolism in the mitochondria of cancer cells. Exploiting this bit of knowledge, Dr. Bingham’s team worked to develop a nonreductive, noncatalytic analog of PDH lipoate, CPI-613. The investigators reasoned that CPI-613 had the potential to attack kinase control of PDH activity, and to do so selectively in tumor cells.
When CPI-613 was evaluated in cell cultures, cells derived from cancer tumors were seen to die when the drug was present at 100 µM. Normal, noncancerous cells remained viable. Moreover, tumor cells in the more hostile in vivo solid tumor environment are even more sensitive to CPI-613.
“Metabolism in cancer cells is different from normal cells,” notes Dr. Bingham. “The changes are predictable. They tend to converge on several metabolic properties. Metabolic drugs (or drugs with a metabolic mechanism of action) can be used to develop cancer therapies effective in many patients, not just small subgroups.
“Many new cancer drugs only target cancers that carry a specific mutation. Metabolic drugs target a metabolic process common to many or all cancers. Hopefully, these new therapies can be used on a broader range of cancers.”
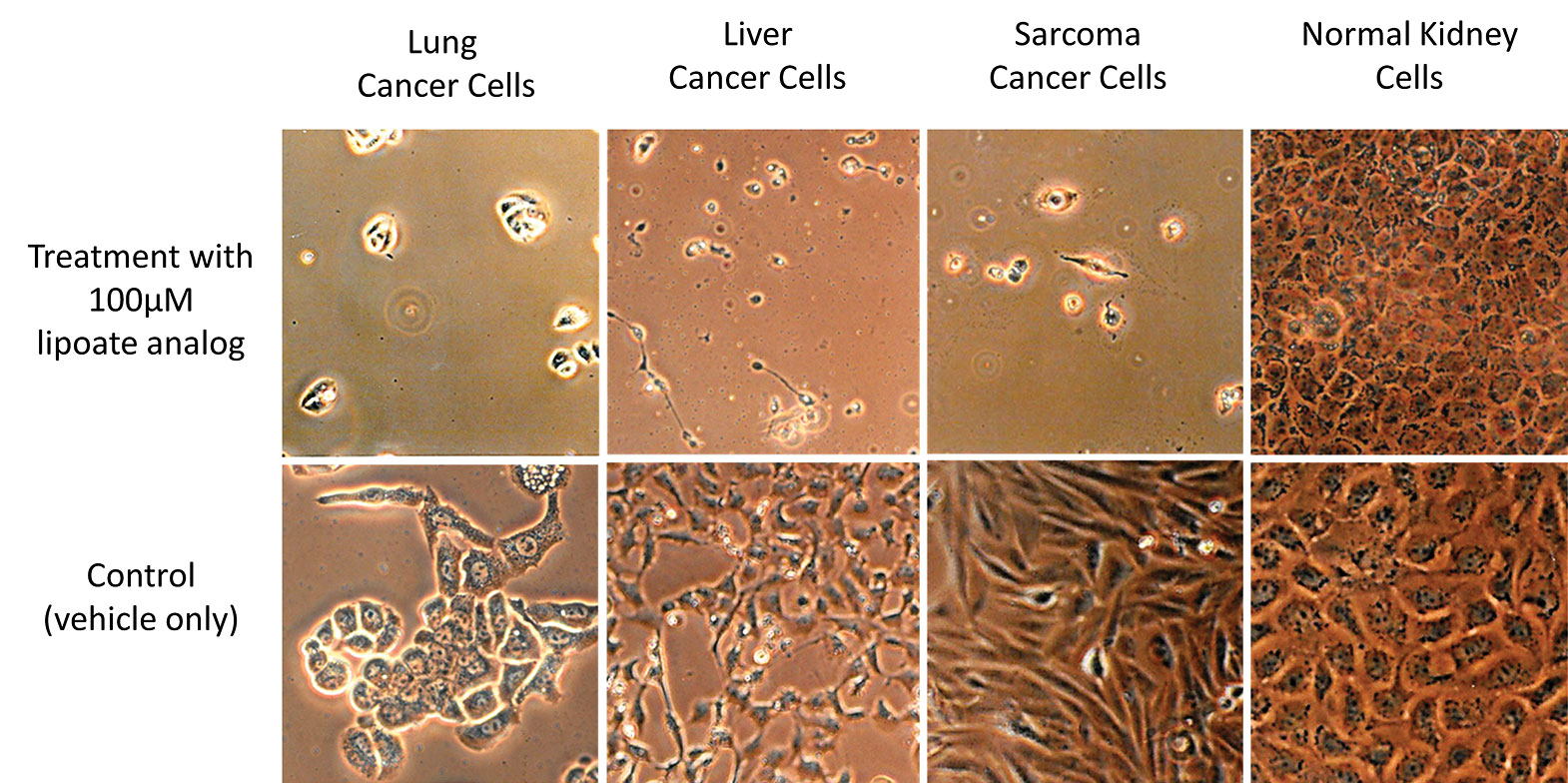
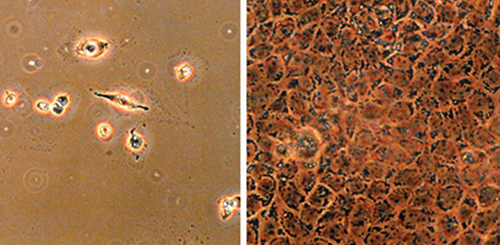
Researchers at Stony Brook University, in the lab of Paul Bingham, Ph.D., found a lipoate analog that can selectively induce inhibition of pyruvate dehydrogenase and a-ketoglutarate dehydrogenase, mitochondrial enzymes involved in cancer cell metabolism in vitro. The compound, CPI-613, is now being developed by Cornerstone Pharmaceuticals. Top row: Cells treated with 100 µM CPI-613. (Cell lines derived from cancer tumors undergo cell death, while cell lines derived from normal tissue remain viable.) Bottom row: The same cells treated with vehicle only.
Disrupting One-Carbon Metabolism
Bringing together the power of genetic and metabolic fields, Gregory Ducker, Ph.D., postdoctoral research fellow in the Rabinowitz Laboratory at Princeton University has found a new way to disrupt one-carbon metabolism. In a nutshell, one-carbon metabolism refers to reactions that move single carbon groups around cellular metabolic pathways.
“One-carbon metabolism plays a big role in the production and methylation of DNA,” explains Dr. Ducker. “Having too much DNA methylation due to an overabundance of one-carbon metabolism is bad for the cell. It can cause hypermethylation (and thus inactivation) of tumor-suppressor genes. We started investigating one-carbon metabolism with a question: Can cancer genetics lead us to new druggable genes in the one-carbon pathway?
“During initial literature searches, we focused on mitochondrial genes that process serine into one-carbon units, because they have been recently shown to be highly upregulated in cancer. We began by targeting the most overexpressed metabolic enzyme in cancer, MTHFD2.”
This enzyme, which processes one-carbon units in the mitochondria, has been cited in several studies as a potential drug target. However, when MTHFD2 gene knockdown was evaluated in cell culture and xenograft models, the results were disappointing. A treatment targeting just this enzyme, it appeared, would not be so effective as to warrant development.
“Most likely, the lack of efficacy was due to cells compensating for the loss of mitochondrial MTHFD2 through activation of the cytosolic one-carbon pathway via SHMT1,” Dr. Ducker suggests. “Knocking out both of these enzymes, however, one mitochondrial and one cytosolic, was lethal in cell culture, but could be fully rescued by media supplementation with formate or thymidine.
“With the knockout of both genes, the ability of cancer cells to utilize serine for its metabolic needs is greatly reduced. When cells with both genes deleted are injected into immune-compromised mice, there is no tumor growth; deleting just one of these genes still allows for tumor development and proliferation. This is surprising because the double-knockout cells would grow in culture, but not as a xenografted tumor.”
“Employing metabolomic profiles of cells as an approach to discover drugs worked well for us,” asserts Dr. Ducker. “We developed these genetic knockouts for different metabolic enzymes, which then can be used to develop a metabolomic fingerprint. Next, we can screen compounds and match the metabolomic fingerprint from the genetic knockout to the desired metabolomic fingerprint induced by the compounds tested, to develop hits based on metabolomic differences.”
Taking Amino Acids off the Menu
Another first-in-class cancer therapy targets amino acid synthesis in cancer cells. “In addition to a higher metabolism of glucose by cancer cells (the Warburg effect), cancer tumors also use amino acids as a nutrient source,” states Francesco Parlati, Ph.D., vice president of research at Calithera Biosciences. Dr. Parlati and colleagues followed up on this observation by developing a small molecule called CB-839. It is a compound that functions as a glutaminase inhibitor. Specifically, it blocks the ability of tumors to use glutamine for metabolic needs.
“We discovered CB-839 at Calithera while looking for compounds which target metabolism,” he recalls. “We knew that tumor cells also consume a lot of glutamine, the most abundant amino acids in the blood plasma of humans. Hence, targeting glutaminase inhibitors made sense. After several rounds of perfecting the candidate via structure–activity relationship (SAR) studies, we saw good results in triple-negative breast cancer cell lines and renal cell carcinoma cell lines.”
“CB-839 showed anticancer activity in several preclinical models,” reports Dr. Parlati. “Mechanistically, CB-839 reduces the levels of key intermediates in the tricarboxylic acid cycle, as well as levels of glutathione and nucleotides. Looking at potential combinations of cancer drugs, we rationalized that using a glutaminase inhibitor (CB-839) with an established drug that inhibits glucose metabolism, like everolimus, would improve the effectiveness of treatment.”
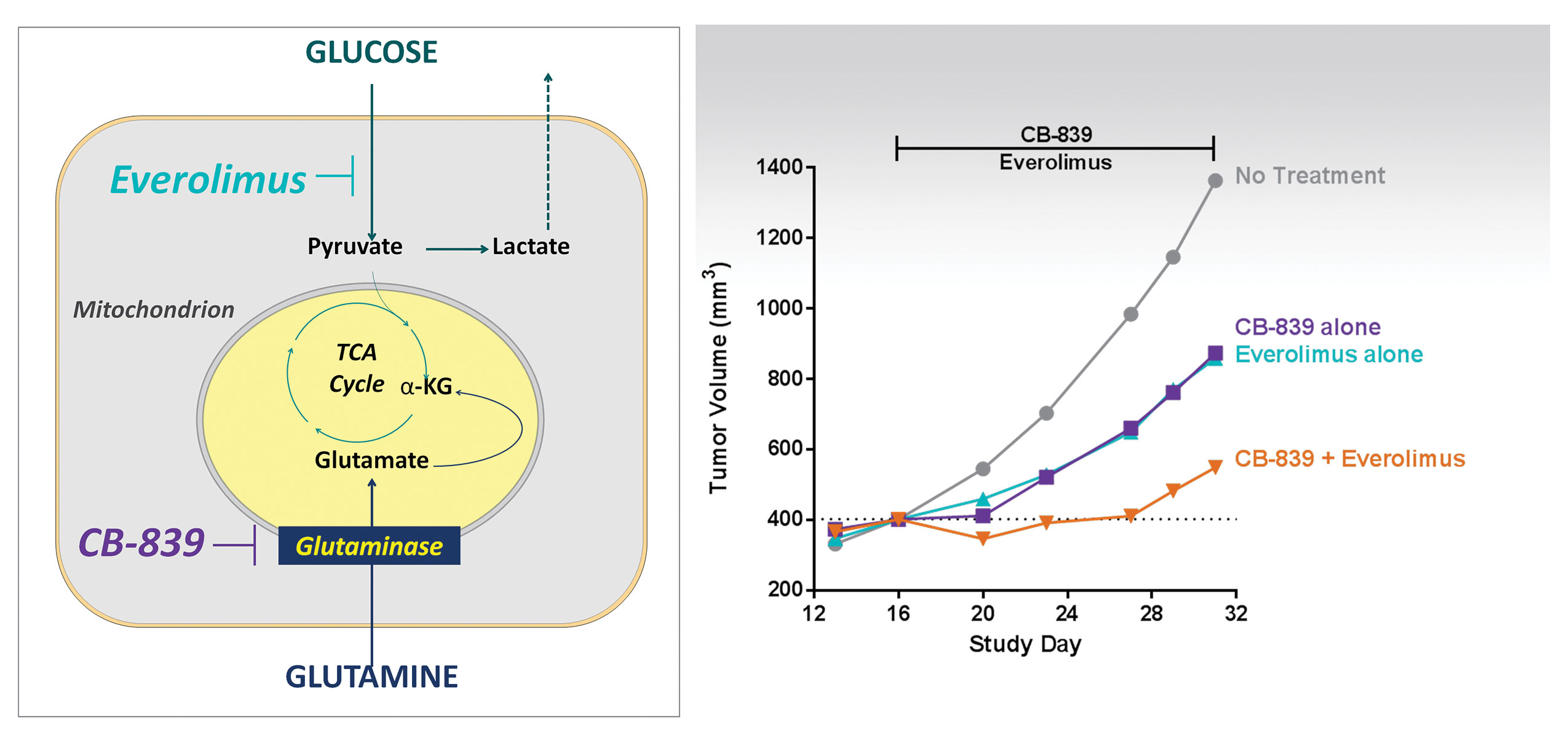
Cancer cells utilize both glucose and glutamine for growth. At Calithera Biosciences, scientists have found that a cancer cell’s ability to use glutamine for growth can be inhibited by using a potent, specific glutaminase inhibitor, CB-839. In combination with everolimus, CB-839 leads to a synergistic effect in decreasing tumor size.
Imaging Metabolic Flux
Another approach to drug development can be found in the imaging of tumors in patients. Magnetic resonance imaging (MRI) allows the visualization of tumor anatomy. However, the metabolism of tumors can also be assessed with a few modifications.
“Normally, MRI depends on the spin of electrons in the protons of water,” says John Kurhanewicz, Ph.D., professor of pharmaceutical chemistry at the University of California, San Francisco. “This is not a problem, as the human body is made up of lots of water. But to look at other molecules, we need to be able to go after different atoms. The simplest way to do this, without getting into complicated physics, it to label compounds of interest with carbon-13 molecules.”
Carbon-13 is a naturally occurring, nonradioactive isotope of carbon; it occurs in about 1% of all the carbon in nature, with 99% of the carbon in natural being carbon-12. The take-home message is that organic compounds labeled with carbon-13 can be detected on MRI imaging.
“Unfortunately, the signal for a carbon-13 labeled compounds is very low,” John Kurhanewicz points out. “To boost the signal, we hyperpolarize the carbon-13 atoms in a glucose molecule immediately before transfusing the solution into patients. This hyperpolarization step boosts the signal intensity about 100,000 times that of nonhyperpolarized solutions.”
Applications of this technique to prostate cancers are especially interesting. The information about glucose metabolism of an abnormal growth of the prostate gland is added to other source of information, such as the anatomical findings from normal MRI, studies of blood supply and perfusion, and microstructure changes. This multiparametric tool is very powerful diagnostically.
In cases of prostate cancer, clinicians must answer a key question: How aggressive is the tumor? The answer will help the clinician develop a course of treatment.
“Having recourse to an approach that exploits carbon-13-labeled compounds, we can determine the pyruvate to lactate flux,” asserts Dr. Kurhanewicz. “This particular flux goes up much higher in high-grade prostate cancers. Hence, we have developed this as a biomarker for aggressive prostate cancer.”
Dr. Kurhanewicz believes that there is much room for growth by applying this technique to other cancers. The monitoring of hyperpolarized carbon-13 pyruvate-to-lactate flux is just one of the technique’s potential applications.
“We have also been able to look at pH and redox levels,” Dr. Kurhanewicz elaborates. “We are interested in collaboratively moving forward to apply this approach to a variety of cancers. This sort of work could be used to aid in the diagnosis and treatment of cancer patients as well as stratifying patients for clinical trials and/or treatments.”
Innovations in technical fields continue to offer promising approaches to finding new treatments for cancer. Additionally, these first-in-class candidates further the understanding of cancer metabolism, by proving where interference with metabolism inhibits the ability of cancer cells to proliferate and survive.