Meghaan M. Ferreira Ph.D. Contributor GEN and Clinical OMICs
From Sturdy Foundations of Structured Data Rises an Imposing Pharmacological Edifice
In 2003, scientists had a lot to celebrate. It was the 50th anniversary of Watson and Crick’s discovery of the DNA double helix. It was also the year scientists witnessed another breakthrough—the sequencing of the human genome. In the glow of the Human Genome Project’s completion, pharmaceutical companies warmed to the notion that the human genome could be read like a set of blueprints. Soon, the designers and builders of the pharmacological world use the blueprints to bring new cures to market.
Over a decade later, drugs approved by the Food and Drug Administration (FDA) target less than 10% of the “druggable genome”—the subset of an estimated 3,000 genes that encode proteins with binding pockets capable of holding small molecule drugs. Much of the druggable genome remains a mystery. As a result, researchers have barely scratched the surface of this potentially rich pharmaceutical resource. Moreover, the Harlow-Knapp effect—the tendency among biomedical and pharmacological researchers to focus their efforts on a small portion of the proteome—suggests that many proteins will remain unstudied until a new biological discovery, such as a disease association or molecular tool, catalyzes their investigation. In other words, many parts of the proteome, and the genome, may remain in the “dark.”
A Light in Dark Places
The National Institutes of Health (NIH) has established the “Illuminating the Druggable Genome” (IDG) program to demystify the dark corners of the druggable genome. The program has decided to focus their initial efforts on four druggable protein families that constitute a significant portion of the proteins expressed by the druggable genome: ion channels, G-protein coupled receptors (GPCRs), nuclear receptors (NRs), and protein kinases.
To increase knowledge surrounding the biochemical properties, biological functions, and disease associations of understudied proteins within these four families, the IDG has developed two complementary projects: the Knowledge Management Center (KMC) and the Technology Development (TechDev) network. The KMC aims to integrate, organize, and disperse information about understudied, druggable proteins. The TechDev network develops technologies and streamlined workflows to ascertain protein function using high-throughput methods amenable to unbiased studies.
Ion channels, one protein family on the IDG’s watch list, are a main source for human disease, and their dysfunction results in a number of channelopathies including epilepsy, diabetes, kidney dysfunction, and heart arrhythmia. However, drugs developed to target ion channels often “don’t work as expected,” remarks Susumu Tomita, Ph.D., professor of cellular and molecular physiology and neuroscience at Yale University School of Medicine. Dr. Tomita notes that the activities of ion channels can be modified if ion channel proteins form complexes with additional proteins in vivo. Most drug discovery platforms don’t sufficiently capture the complicated architecture of these in vivo complexes.
Using a high-throughput method, Dr. Tomita’s lab is systematically screening every human gene—or at least the 17,000 distinct gene constructs currently stored in the lab’s freezer—to identify proteins that regulate ion channel function. The lab uses a FLIPR® assay with fluorescent dyes sensitive to calcium, potassium, and membrane potential to measure ion channel activity in HEK (human embryonic kidney) cells cultured in 384-well plates and transfected with the gene of interest. According to Dr. Tomita, his team was the first to use this approach to identify functional modulators for genes.
The lab’s approach has already yielded interesting findings, such as those described in an article that appeared in the March 2017 issue of Neuron. This article describes how Dr. Tomita’s team established a molecular mechanism for ion channel localization to the synapse, an essential step in neuronal signaling and brain function, through interactions with GARLHs—subunits of GABAA receptors consisting of LH4 and LH3 proteins.

Small Models, Big Plans
Dr. Tomita’s lab has also collaborated with IDG investigators David Kokel, Ph.D., assistant professor of physiology at the University of California, San Francisco; Joanna Yeh, Ph.D., assistant professor of medicine at Harvard Medical School; and Randall Peterson, Ph.D., professor of pharmacology and toxicology at the University of Utah. The collaboration has produced a unique animal model for childhood absence epilepsy by knocking out the CACNG3 gene in zebrafish, which produced a seizure phenotype reminiscent of that observed in humans with CACNG3 mutations.
Zebrafish larvae, which are virtually microscopic, provide an inexpensive, scalable model that can, when combined with genomic engineering tools, create a powerful disease-modeling platform. Also, these tiny tropical fish, by dint of their aquatic nature, are easy to dose with drug compounds. Thus, zebrafish models make it possible to screen for drugs, such as anticonvulsants, in a high-throughput manner not possible with other animal models.
Although they are still preliminary, the results of this collaboration testify to the big impact a little zebrafish can have on our approach to chemical biology. “By making genetic knockdowns in poorly understood genes,” says Dr. Kokel, “we can start to get an idea of how they affect behavior and development.”
Both their transparency and their genetic malleability make zebrafish highly compatible with microscopy and fluorescent reporter systems. Interest in exploiting the zebrafish model’s reporter compatibility has led to another IDG collaboration, an effort involving Steve Finkbeiner, M.D., Ph.D., the director of the Taube/Koret Center for Neurodegenerative Disease Research and a senior researcher at the Gladstone Gladstone Institute of Neurological Disease.
The Finkbeiner lab has developed two key technologies: a robotic fluorescent microscope developed for longitudinal studies, and a toolbox of fluorescent biosensors for different cellular functions. The lab uses these tools to build human, preclinical models for neurodegenerative diseases, such as Parkinson’s disease.
The flexibility of the lab’s platform allows researchers to move seamlessly from pathway screening to drug target identification by introducing different genes into their models, performing a “physical exam” on cultured induced pluripotent stem cell (iPSC)-derived neurons, and monitoring cellular disease characteristics—such as protein aggregation or mitochondrial disruption—to determine if the gene rescued or exacerbated the disease phenotype. “Similar to doing a clinical trial in patients, you can then ask what role those phenotypes play in the degenerative process,” explains Gaia Skibinski, Ph.D., a staff research scientist at Gladstone.
For the IDG project, the Finkbeiner lab has developed a new biosensor that indirectly measures GPCR activity by quantifying the migration of the downstream transcription factor NFAT (nuclear factor of activated T cells) as it moves from the cytosol to the nucleus upon elevation of intracellular calcium levels and GPCR activation.
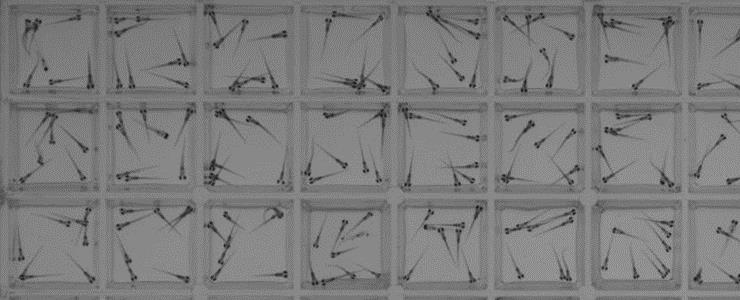
Representative image of a 96-well plate fully loaded with 8-10 animals per well. The zebrafish are 7 days old. [David Kokel/Joanna Yeh]
Filling Out Logical Frameworks
As the Finkbeiner Lab and other TechDev groups discover the pages missing from the druggable genome story, the KMC will collate them, along with existing knowledge, to create a single, searchable, web-based informatics site—Pharos. Researchers worldwide can then use the information to prioritize and inform their own experimental objectives.
“[It] may seem a little bit backwards,” comments Lars Juhl Jensen, Ph.D., professor of cellular network biology at the Novo Nordisk Foundation Center for Protein Research, University of Copenhagen. “The way to find out which proteins we lack knowledge about is by collecting the knowledge we have and then basically seeing where the hole is.”
Dr. Jensen has helped the IDG classify the developmental level of individual genes and proteins based on how frequently they appear in biomedical, funding, and patent literature using a text-mining technique called named entity recognition. “A lot of the data that’s out there is available only in the form of text, which, from a bioinformatician stand point, is the most inconvenient way you could possibly give me the data,” explains Dr. Jensen. Text mining extracts the information from text, and translates it into a more palatable form for data analysis.
In addition to aiding in the classification of proteins based on their abundance (or paucity) in literature, Dr. Jensen has also contributed extensive genome-wide datasets for tissue expression and gene-disease relationships that could motivate research and funding for understudied genes and proteins.
Text mining provides both a necessary and powerful tool for curating scientific data, but the KMC has also made organizing data in a meaningful way a top priority. To provide an organizational framework for drug target information, the IDG has arranged for Pharos to integrate the Drug Target Ontology (DTO), a classification framework developed by Stephan C. Schürer, Ph.D., associate professor of molecular and cellular pharmacology at the University of Miami.
“The challenge in biology is often incomplete knowledge,” states Dr. Schürer, who asserts that organizing data through an ontology creates a flexible structure that can accommodate new scientific discoveries and conflicts in data more readily than traditional database models.
The DTO interface provides a visual representation of the hierarchical structure of the ontology, which classifies drug targets according to their protein family, binding ligands, disease relevance, and tissue expression. “The overall goal,” comments Dr. Schürer, “is to provide a better way for researchers to identify or evaluate targets of interest.”
General inquiries about a disease, binding ligand, or tissue highlights associated proteins (color coded according to their developmental level) within circular boundaries that represent the protein family or subfamily to which they belong, whereas specific queries link to a detailed summary of information collected for individual proteins.
Ontologies can also infer information that is not explicitly given, by creating logical definitions that relate different pieces of information to one another. Also, according to Avi Ma’ayan, Ph.D., professor of pharmacological sciences at the Icahn School of Medicine at Mount Sinai, a lot more is known about understudied genes and proteins than we may think. “A lot of information,” Dr. Ma’ayan insists, “can be inferred, organized, and extracted using publicly available ‘omics’ datasets.”
The Ma’ayan laboratory has used this modus operandi to develop a searchable web-based relational database called Harmonizome to provide a comprehensive knowledge resource for genes and proteins. Currently, Dr. Ma’ayan plans to expand the capabilities of Harmonizome, which the IDG has integrated into Pharos, by including a feature that can make predictions about gene function based on co-expression with other genes.
“If [understudied proteins] are correlated with known proteins, we can infer their function and figure out where they are in the cell, what they do, and what other [protein] interactions they have,” Dr. Ma’ayan explains.
“The [IDG] program itself is helpful in changing the spirit of the field,” Dr. Ma’ayan continues. He adds that the shift has arrived during a genomics and proteomics revolution that has produced more powerful technology for studying the druggable genome.
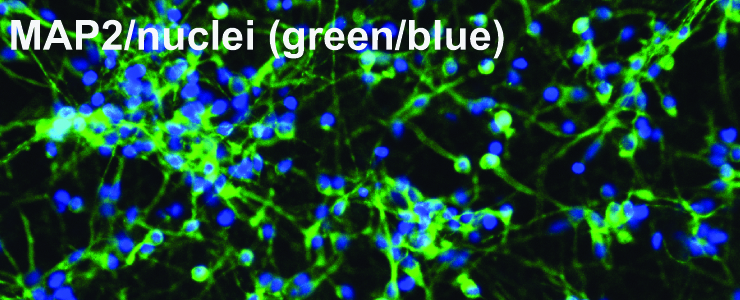
Representative image from immunocytochemistry of human neurons differentiated from iPSCs from a control individual, co-stained with MAP2 and DAPI. Approximately 75% of cells are MAP2 positive. Scale bar is 40 µm. Skibinski G., Hwang V., Ando D.M. et al (2017), PNAS. doi: 10.1073/pnas.1522872114]
Genomic Renderings
Architectural notions of the sort mentioned in this article’s introduction were entertained by the authors of “A Vision for the Future of Genomics Research,” an article published in Nature following the completion of the Human Genome Project. These authors, writing on behalf of the U.S. National Human Genome Research Institute, characterized the Human Genome Project as a “firm bedrock foundation” upon which a noble structure, a new phase of research that would realize the “true promise of genomics for benefitting humankind.”
Like any construction that would reach a great height, this noble structure requires an exceptionally solid foundation—one that is still being reinforced. While this work proceeds, the architectural renderings of what is yet to be built are as inspiring as ever—as they should be.
Here, let us follow the authors of the “Vision” article by quoting the architect Daniel Burnham: “Make no little plans; they have no magic to stir men's blood and probably will themselves not be realized. Make big plans; aim high in hope and work, remembering that a noble, logical diagram once recorded will not die, but long after we are gone will be a living thing, asserting itself with ever-growing insistency.”







