Optimization of experimental assay design is crucial for the overall success of any drug discovery initiative. The factors that need to be considered include: selection of the most appropriate assay technologies, reading modalities, and experimental models (biochemical, cellular, or animal). Collectively, these factors are known to influence data quality, biological relevance, and therapeutic predictability, which will ultimately impact the success of entire preclinical drug development efforts.
Assay technology selection
Biological relevance and therapeutic predictability
The first step in selecting an assay technology is to question the paradigms to be addressed. Some examples are:
- Activation, inhibition, or modulation of a target
- Elucidation of MOA (mechanism of action)
- Determination of binding affinity of receptor–ligand or protein–protein interactions
- Identification and quantitation of disease-specific biomarker or biomarker panel in plasma
Defining the paradigms and predictable end point(s) enables selection of the assay formats and technologies to be used for the study.
The assay technology selected needs to reproduce physiologically relevant conditions; that is, it needs to be performed in relevant in vitro systems with pharmacologically relevant concentrations of components to yield biologically relevant data. Phenotypic cellular assay models more accurately reflect the complex biology of disease, allows for multiparametric measurement of ex vivo or in vivo events, and enables the characterization of intractable target(s).
However, such assays sometimes require complex execution, which can impede the screening process. Biochemical assays are simpler to execute than their cellular counterparts and offer better consistency. Unfortunately, the latter don’t guarantee high physiological relevance. Hence, it is desirable to start with a biochemical assay followed by a cell-based assay to corroborate results in a physiological context (that is, to provide orthogonal validation).
Sample matrix compatibility
Biomarker assays aim to detect or quantitate targets in biological samples (for example, complex matrices) ranging from secreted proteins in cell culture supernatants to cell lysates, tissue extracts, saliva, urine, serum, plasma, blood, or other fluids (for example, CSF, BALF). Assay technologies may not always be compatible with particular matrices, which can cause interference. For example, hemoglobin found in serum, plasma, or blood exhibits wide absorbance across the visible light spectrum, namely between 350 and 600 nm. Assay technologies relying on excitation or emission at these wavelengths will be prone to interference. Wash-based assays can be used to circumvent interference caused by highly complex sample types, as molecules within the matrix that contribute to interference are washed away. To determine sample matrix compatibility, spike-and-recovery and linearity experiments are traditionally performed in the appropriate diluent for standard curves.
Additionally, for complex biological matrices, antibody selection can be challenging since protein targets secreted from cells into cell culture media or serum may be cleaved or modified, altering epitope recognition—hence the benefit of using protease cocktail inhibitors.
Assay performance
Sensitivity
Sensitivity of an assay dictates the detection of the target within the dynamic range of the assay with the caveat that the lower limit of detection could vary marginally from batch to batch.
Sensitivity is also dependent on sample type. Physiological levels of an analyte in serum differ from the levels secreted by a given cell line. “Precious” samples, such as limited cells or tissue, impact the permissible volume for testing. Technologically, luminescent and fluorescent modes offer greater sensitivity compared to absorbance assays. Both assay format and detection technology play major roles in sensitivity.
Dynamic range
The dynamic range of an assay defines both the lower and upper limits of detection. For example, as shown in Figure 1A, the dynamic range for quantitating tumor necrosis factor-α (TNFα) varies by at least a log across different technologies. Furthermore, measurable binding affinities for various analyte interactions can vary based on the chemistry and detection technology (Figure 1B). Therefore, when samples fall outside the limits of detection of an assay, concentrations of analyte, kinetic parameters, potencies, and binding affinities could result in variations of measurement. Hence, cell or analyte titrations and calibration curves are implemented to ensure detection windows and avoid saturation.
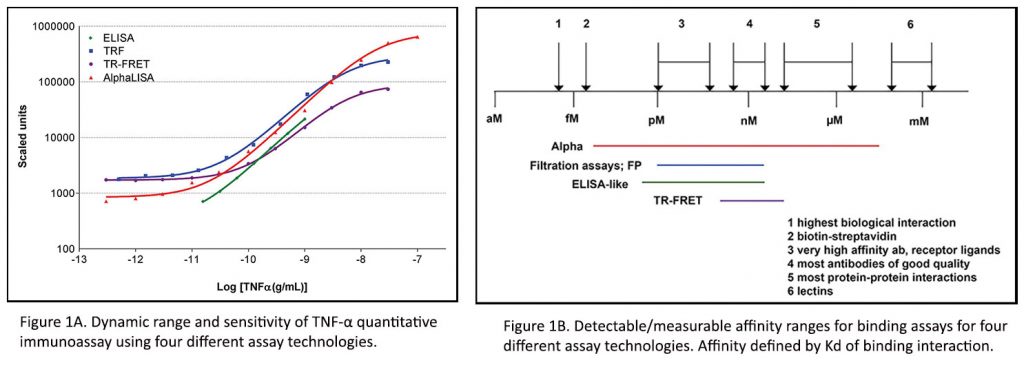
Specificity
Specificity is important to ensure that the assay will measure the desired target or phenotype. For immunoassays, the appropriate antibody-to-target-analyte specificity needs to be considered especially since targets may vary from modified moiety or cleaved form (neoepitope) of a protein to bound/unbound or active/inactive forms. In addition, species and target cross-reactivity need to be considered.
For assays involving conjugation or labeling of antibodies, specificity should be confirmed. For biochemical enzymatic assays, substrate selection is important to achieve specificity.
For example, many recombinant, purified protein kinases can be used with generic or specific peptides, or whole protein substrates. In addition, to prove specificity, reference inhibitors should be validated for the developed assay.
Robustness and accuracy
Although signal intensity or signal-to-background ratio (S/B) of an assay is typically evaluated, it is vital that the assay is robust enough to yield highly reproducible and accurate data. For instance, the Z-prime value (Z‘) is usually a better indicator of robustness compared to S/B. For example, a low signal intensity (low S/B) but a high Z‘ assay is better than a high signal (high S/B) but a low Z‘. Also, the assay should be highly reproducible, delivering the same quality data irrespective of the day or the individual running the assay. Quantitative assays should be calibrated against an appropriate standard. Reliability of an assay can be achieved using internal and external controls and an array of reference calibrators, antibodies, and compounds as appropriate.
Semiquantitative or qualitative assays should be run using reference compounds or binding molecules as appropriate. Acceptable levels of robustness, reproducibility, and accuracy are dictated by the goals of the assay and must be determined by the researcher.
Reagents and consumables
Reagent quality
For immunoassays, one must consider the selection of quality antibodies based on their sensitivity, specificity, and continuous availability (that is, reproducible production). Relying on monoclonal antibodies (mAbs) guarantees the latter aspect. Similarly, validated high-quality enzymes and recombinant proteins should be used for developing biochemical enzymatic and binding assays.
Reagent titration and optimization
In general, all reagents (such as cells, proteins, 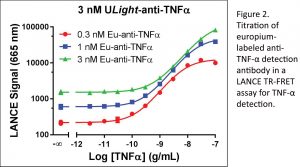 enzymes, cofactors, substrates, stimulators, inhibitors, agonists, and antagonists) utilized in an assay need to be titrated in cross-titrations or stepwise via optimal concentrations. The process ensures steady-state kinetics and prevents saturating concentrations to maximize S/B while reducing reagent consumption (Figure 2).
enzymes, cofactors, substrates, stimulators, inhibitors, agonists, and antagonists) utilized in an assay need to be titrated in cross-titrations or stepwise via optimal concentrations. The process ensures steady-state kinetics and prevents saturating concentrations to maximize S/B while reducing reagent consumption (Figure 2).
Cell models
When choosing cellular models, one should consider the advantages and disadvantages of using primary cells vs. recombinant/immortalized cell lines, and the study of endogenous vs. recombinantly expressed target proteins. The target expression level needs to be assessed, as too much or too little expression can affect sensitivity and assay quality. Cell passaging and culture conditions can also affect functional response.
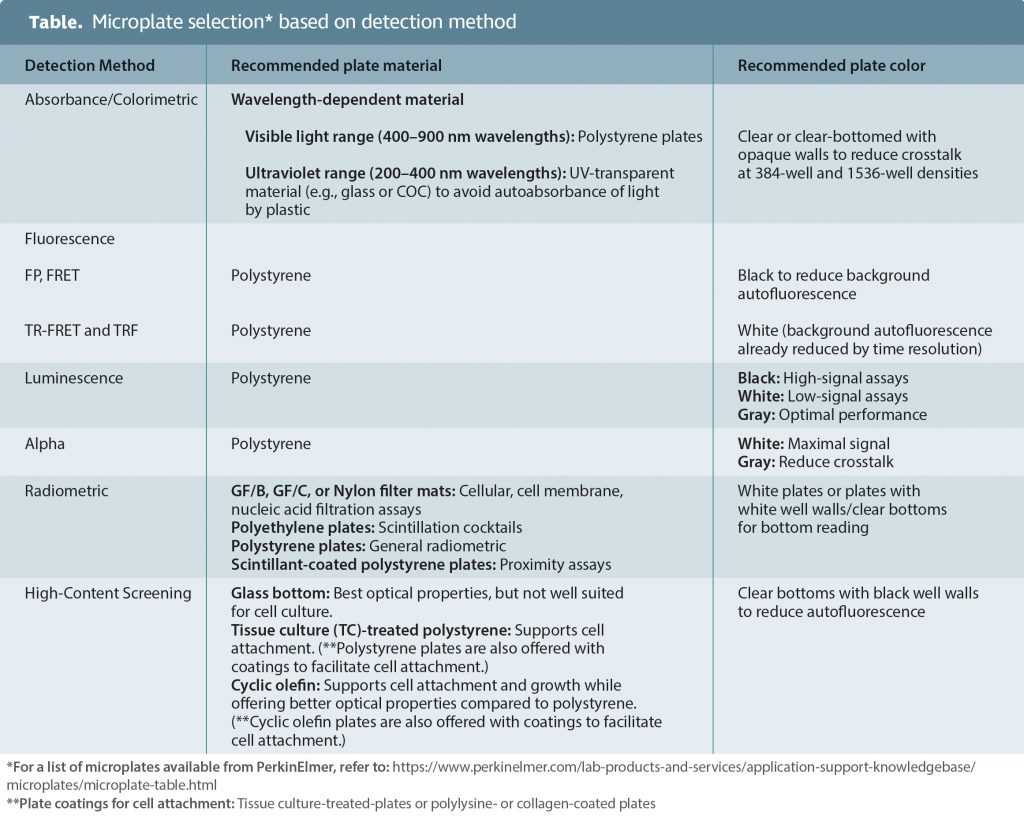
Microplate selection
Correct selection of a microplate can improve assay performance by reducing crosstalk, decreasing background, minimizing signal absorbance, and/or allowing for signal amplification. The Table shows a matrix for microplate selection based on the method of detection.
Summary
Drug discovery initiatives are known to be costly and time consuming. Optimizing experimental factors known to influence data quality, biological relevance, and therapeutic predictability ultimately drives the success of entire preclinical drug development efforts.
Selecting the most appropriate assay technologies, reading modalities, and experimental models are examples of experimental factors to optimize for laying the groundwork of successful and cost-effective drug development.
Catherine Lautenschlager, PhD, is e-marketing manager, Roger Bosse, PhD, is sales development leader, Jen Carlstrom, PhD, is senior applications scientist, and Anis H. Khimani, PhD, is director, strategy leader and applications, for PerkinElmer’s Discovery & Analytical Solutions business.