![This array produced from a software program called Iterative Clustering and Guide-Gene Selection shows the expression of genes (horizontal rows) that are associated with different types of bone marrow progenitor cells (vertical columns). It gives scientists an unbiased way to identify developing cells in various intermediate states and discover new cellular intermediates. Yellow indicates highly expressed genes in particular cells, whereas blue indicates low or no expression. [Cincinnati Children's]](https://genengnews.com/wp-content/uploads/2018/08/Sep1_2016_CincinnatiChildrens_Genes1706018813-1.jpg)
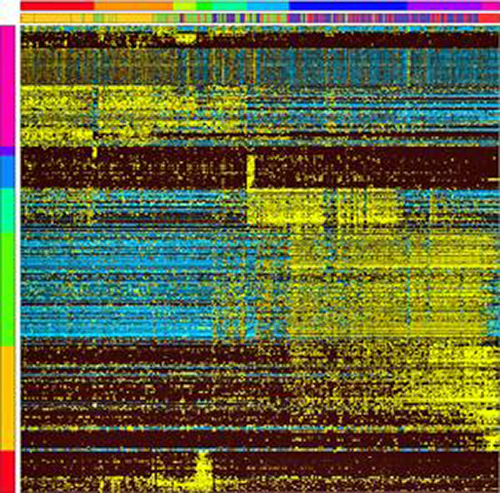
This array produced from a software program called Iterative Clustering and Guide-Gene Selection shows the expression of genes (horizontal rows) that are associated with different types of bone marrow progenitor cells (vertical columns). It gives scientists an unbiased way to identify developing cells in various intermediate states and discover new cellular intermediates. Yellow indicates highly expressed genes in particular cells, whereas blue indicates low or no expression. [Cincinnati Children’s]
The sage who said, “When you come to a fork in the road, take it,” wasn’t thinking of cell development, but his words aptly describe the confusion that comes over progenitor cells. For example, there are progenitor cells that are known to occupy bipotential intermediate states, manifesting mixed-lineage patterns of gene expression. While these states may seem chaotic, they ultimately lead to order. And now, according to a recently published study, there is evidence that cell-fate indecision is more purposeful than anyone might have guessed.
Researchers at Cincinnati Children's Hospital Medical Center report that as developing blood cells are triggered by a multitude of genetic signals firing on and off, they are pulled back and forth in fluctuating multilineage states before finally becoming specific cell types. Although the scientists still don't understand exactly what cues the cells to an eventual fate, their work suggests that competing gene networks induce dynamic instability, resulting in mixed-lineage states that are necessary to prime newly forming cells for that decision.
The findings appeared August 31 in the journal Nature, in an article entitled, “Single-Cell Analysis of Mixed-Lineage States Leading to a Cinary Cell Fate Choice.” The article explores the possibility that mixed-lineage states may reflect the molecular priming of different developmental potentials by co-expressed alternative-lineage determinants, namely transcription factors.
To investigate mixed-lineage states, a team of scientists led by Cincinnati Children’s Harinder Singh, Ph.D., and H. Leighton Grimes, Ph.D., used single-cell RNA sequencing coupled with a new analytic tool, iterative clustering and guide-gene selection, as well as clonogenic assays to delineate hierarchical genomic and regulatory states that culminate in neutrophil or macrophage specification in mice.
“We show that this analysis captured prevalent mixed-lineage intermediates that manifested concurrent expression of haematopoietic stem cell/progenitor and myeloid progenitor cell genes,” wrote the authors of the Nature article. “It also revealed rare metastable intermediates that had collapsed the haematopoietic stem cell/progenitor gene expression programme, instead expressing low levels of the myeloid determinants, Irf8 and Gfi1.”
Prior to the current study it already had been proposed that neutrophil and macrophage blood cells result from a bistable gene regulatory network (one that can manifest either of two stable states). But the different cellular transition states and underlying molecular dynamics of development have remained unknown.
Repeatedly working back and forth between laboratory biology and computational analysis, authors of the current paper say their analysis of developing blood cells captured a prevalent mixed-lineage intermediate. These intermediates expressed a combination of genes, including those typical of stem cells that can give rise to all blood cell types as well as some genes that are specific for red blood cells, platelets, macrophages, and neutrophils. This seemed to reflect competing genetic programs.
The research team also found the developing cells moving through a rare state where they encountered turbulence, termed “dynamic instability.” This was seen to be caused by two counteracting myeloid gene regulatory networks. Two key components of the counteracting gene networks were Irf8 and Gfi1, genes that are involved in blood cell formation. Both genes encode transcription factors that control what genetic information is used in the formation of a cell. When Irf8 and Gfi1 were eliminated from the picture, researchers showed that the rare cells could be trapped in an intermediate state.
“It is somewhat chaotic, but from that chaos results order,” said Dr. Singh. “It's a finding that helps us address a fundamental question of developmental biology—what is the nature of the intermediate states and the networks of regulatory genes that underlie cell-type specification.”
As they continue their research, the authors want a clearer understanding of what finally causes cells in intermediate states of dynamic instability to assume specific fates. They suggest that the influence of two simultaneous and counteracting gene networks generates internal oscillations that are eventually stabilized by unknown mechanisms to generate one of two different cell fates.