November 15, 2007 (Vol. 27, No. 20)
Fiona Coats Ph.D.
James Lazar Ph.D.
Sherry Challberg Ph.D.
Bridging the Gap Between Cytoplasmic Signaling and Gene Expression
Marligen Biosciences’ (www.marligen.com) multiplex transcription factor profiling tools have been successfully used to address a variety of problems inherent in drug discovery and development. Applications have ranged from defining mechanisms of disease to identifying drug repositioning opportunities.
Transcription factor (TF) profiling quantitatively and simultaneously measures changes in the DNA binding characteristics of multiple transcription factor complexes isolated from samples. Methods that examine only one or a few such interactions without the context provided by multiplexed TF profiling can only confirm assumptions supporting hypothesis-driven research.
TF profiles provide broad coverage of proteins, pathways, receptors, disease states, and transcriptional activity within cells and tissues. The approach is hypothesis generating rather than hypothesis driven and therefore is a novel and powerful tool for drug research. TF profiling also holds promise for identifying predictive markers of potential toxicity.
Cytoplasmic signaling culminates in the modulation of gene expression through the DNA-binding activity of transcription factors. Since upstream signaling events lead to the activation of a relatively limited number of transcription factors that then regulate expression of many different genes, transcription factor profiles can monitor upstream signaling and predict downstream gene expression. Changes observed in the activity of one or more transcription factors therefore provide signatures of biological responses or disease states.
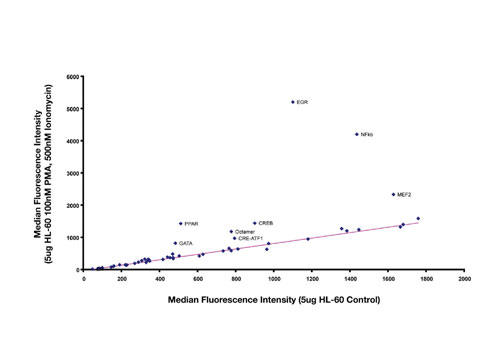
Fig 1 Part 1
Profiling Assay
Marligen’s 50-plex TF profiling assay is a biosensor for over 10,000 upstream signaling events involving over 2,200 proteins. One or more transcription factor activities detected by the 50-plex assay are represented in 75% of established signaling pathways and over 400 different diseases. Strong homologies of binding sites across multiple species means the same panel can be used for analyzing cells and tissues from animal species commonly used in preclinical research, which allows the 50-plex panel to provide coverage of a broad range of biological responses and processes.
Marligen’s TF profiling technology is an ideal precedent for ChIP assays, which are used to identify transcription factor binding sites of interest, by guiding antibody selection to pick the most meaningful and cost-effective ChIP analysis.
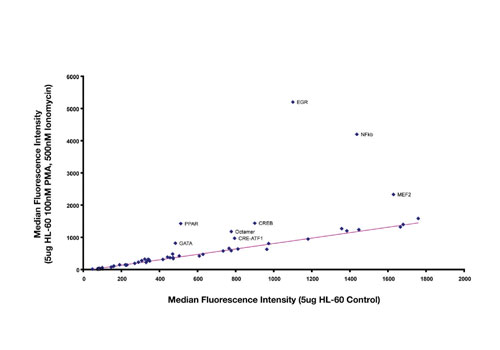
Figure 1 illustrates how results are analyzed to identify changes in the activity of transcription factor complexes isolated from nuclear extracts. In this example, extracts were isolated from HL-60 cells, with or without treatment with 100 nM PMA and 500 nM Ionomycin, and tested using the 50-plex TF profiling assay.
Results from control samples are plotted on the X-axis, and treated samples are plotted on the Y-axis. Each point on the graph represents a different assay within the 50-plex. Strong upregulation was observed with EGR, NFkB, and MEF2, and more modest upregulation was detected for CRE-ATF, CREB, GATA, Octamer, and PPAR. The activities of the remaining TF complexes were unchanged. The near-perfect linearity of the data for binding assays between controls and treated serves as a basis for internal sample normalization.
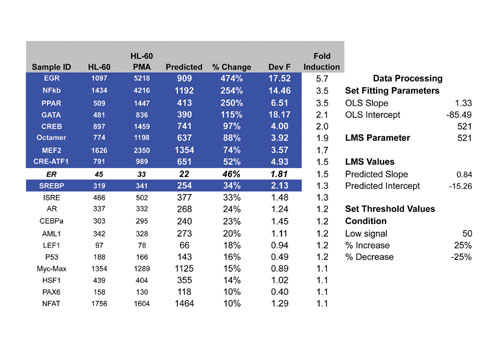
Figure 1 Part 2
The Technology
Samples prepared with Marligen’s nuclear extraction protocol contain activated transcription factor complexes that provide a real-time snapshot of transcriptional regulation. The cytoplasmic fraction can be retained and analyzed separately if desired.
The multiplexed assays employ DNA probes composed of a double-stranded binding site, a biotin molecule for detection, and a unique single-stranded DNA capture sequence. Nuclear extract, typically 5 µg, is incubated with a mixture of the 50 different DNA probes. The TF complexes in the extract bind to their probe-specific cognate recognition sequences in the double-stranded regions of the DNA probes. This mixture is incubated with a digestion reagent that destroys probes that do not contain transcription factor complexes bound to the probe, while probes with bound complexes are protected from digestion.
The probes are captured onto the surfaces of Luminex’(www.luminex.com) XMap® beads via hybridization of single-stranded capture sequences with complementary sequences linked to the surface of the beads. Streptavidin-PE (SA-PE) is added to the mixture and both the identity of the bead and the amount of fluorescence from SA-PE bound to biotin on the DNA probes is measured using the Luminex System.
The accuracy and specificity of the binding sequences are validated using a variety of methods including comparison to gel shifts, assays with purified recombinant proteins and cells transfected with TF expression constructs, comparing observed changes in TF profiles with published results, and competition and pull-down assays.
Reproducibility was extensively validated in collaboration with external pharma researchers, and the CVs for biological replicates were generally less than 10% for each binding site. Marligen’s nuclear extraction protocols and multiplexed assays have been shown to be compatible with many cell types of different origin as well as a wide range of animal tissues.
Applications of TF Profiling
In one study, a researcher had previously tried to understand the underlying disease mechanisms in acute lung injury (ALI) by characterizing differences between normal cells and the ALI model system using high-density gene expression microarrays.
The results from these studies identified almost 2,000 differences between control and treated cells, and the complexity of the data made interpretation a daunting and virtually impossible task. Marligen’s Multiplex Transcription Factor Profiling uncovered the involvement of several transcription factor binding events that were previously unknown, and the technology facilitated kinetic analysis of the changes (Figure 2).
This information resulted in a more focused interpretation of the gene expression array data, identified new regulatory pathways in disease pathogenesis, and may lead to new approaches for developing therapies.
A second study was performed by a pharmaceutical researcher who was trying to understand the poor efficacy of a lead compound observed in a Phase II trial. TF profiles of tissues from animal models of the disease led to an explanation of the failure. It also led to the identification of several metabolic enzymes involved in pathogenesis that represent attractive new therapeutic targets. Finally, a repositioning opportunity for the failed drug in another area was uncovered. As an illustration of off-target effects identified by TF profiling, Marligen scientists performed a 50-plex screen using a known PPAR inhibitor (Figure 3).
TF profiling offers researchers the ability to screen across a broad range of upstream signaling pathways and downstream gene expression involved in disease pathology and drug action. TF profiling has been shown to reveal novel biological effects, define mechanisms of disease and drug action, identify new drug targets, evaluate specificity, and uncover repositioning opportunities. Pharmaceutical and biotech customers are now using the technology in preclinical R&D to understand diseases and more comprehensively screen specificity, thus enabling additional decision points early in pipeline development.
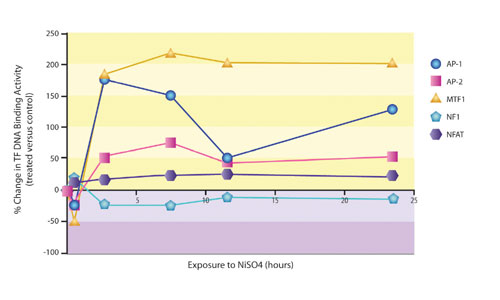
Figure 2
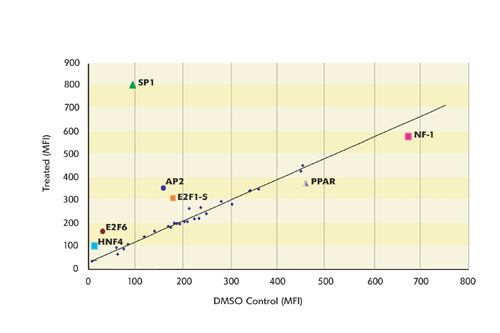
Figure 3
Fiona Coats, Ph.D., is vp, business development, James Lazar, Ph.D., is vp and CSO, and Sherry Challberg, Ph.D., is president and CEO at Marligen Biosciences.
Web: www.marligen.com. Phone: (301) 874-4990. E-mail: [email protected].