August 1, 2018 (Vol. 38, No. 14)
Brian Wang Ph.D. genomic tools market development manager Integrated DNA Technologies
Integrated DNA Technologies Suggests Strategies to Boost Efficiency, Avoid Blunt-End Incorporation, and Reduce Toxicity
In recent years, CRISPR-Cas9 genome editing has enjoyed spectacular success because it combines flexibility and usability. The Cas9 nuclease need only be loaded with a guide RNA to direct it to almost any genomic location, where it will create a double-stranded break that may serve as an insertion point for exogenous DNA. After a cell’s genomic DNA is cut, the cell’s DNA repair mechanisms will repair the break, splicing together the loose ends, whether donor DNA is present or not. If donor DNA is present, the repair may incorporate the exogenous sequence between the (formerly) loose ends.
To carry out the repair process, eukaryotic cells have two mechanisms at their disposal:
- Nonhomologous end joining (NHEJ) can repair a double-stranded break without requiring homologous sequences on both sides of the break. This makes NHEJ a more straightforward form of repair, but it also increases the risk of errors, such as insertions or deletions, at the cut site.
- Homology-directed repair (HDR), or what has been called “true genome editing,” relies on the presence of a template with DNA “arms” homologous to each end of the break. These sequences are used to guide repair and can fix breaks with a vastly reduced error rate.
Due to its greater reliability in DNA repair, HDR is the preferred mechanism if CRISPR-Cas9 is used to insert (or knock-in) exogenous DNA sequences, such as selectable markers or fluorescent tags. Therefore, improving the rate of HDR is a key challenge in achieving reliable insertions. In this tutorial, we describe a series of strategies for obtaining higher rates of HDR based on experimental data from our detailed CRISPR optimization studies.
Single- or Double-Stranded DNA?
Although most researchers use double-stranded DNA, both single- and double-stranded DNA templates are suitable for CRISPR-mediated insertion. However, using a double-stranded template is more likely to result in blunt-end incorporation by NHEJ with duplication of the homology arms.
To reduce the risk of blunt-end incorporation, and thus achieve a higher rate of template incorporation by HDR, single-stranded oligo deoxynucleotides (ssODNs) can be used. Single-stranded templates can be created that link up to 2000 nucleotides and enable high HDR rates, even when short homology arms are used. An additional advantage of ssODNs is that they can be used after standard desalting purification and do not require polyacrylamide gel electrophoresis purification to improve HDR efficiency (Figure 1).
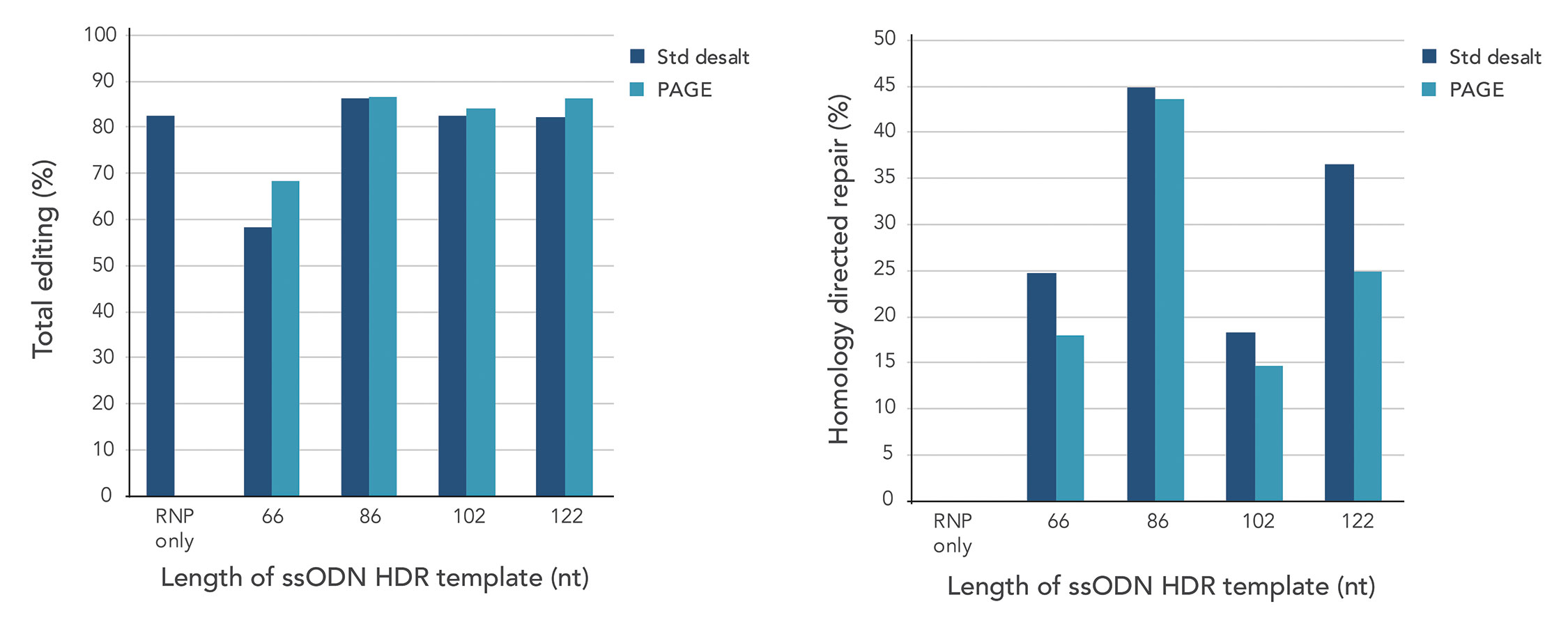
Figure 1. Amplicon sequencing shows high rates of insertion of Ultramer ssODN template (top), with up to 50% of insertions being repaired by HDR (bottom). PAGE purification does not appear to improve insertion compared to standard desalting purification.
Designing an ssODN Template
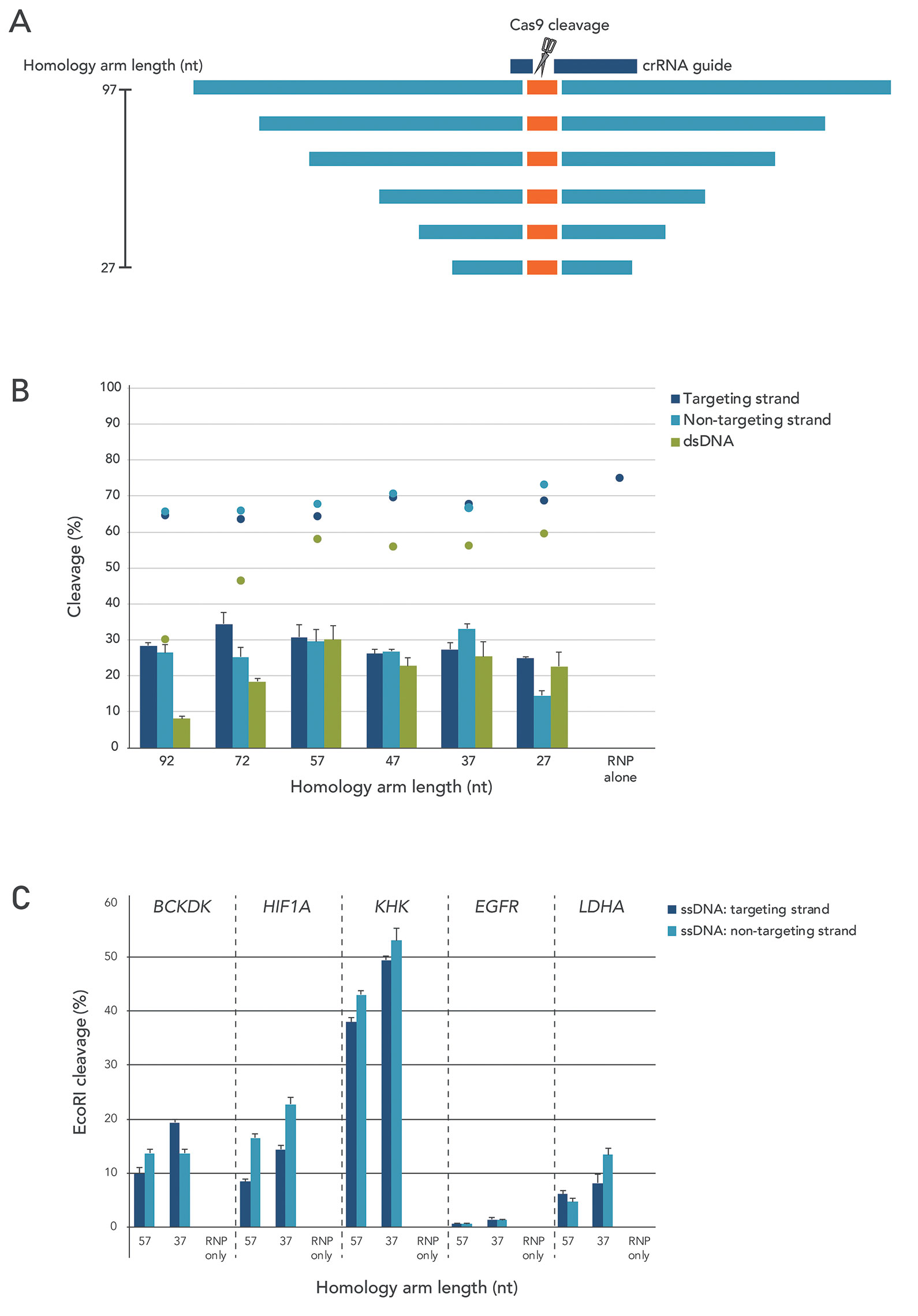
The design of the homology arms is a key factor in stimulating the use of the HDR pathway over NHEJ. To determine how arm length affects template incorporation, we produced a series of templates with arm lengths varying between 27 and 92 nucleotides (Figures 2A & 2B). We also looked at total template incorporation and HDR-mediated incorporation. When creating small insertions using Ultramer® Oligonucleotides (a total maximum donor length of 200 bases), we found good, consistent incorporation at arm lengths between 30 and 60 nucleotides
When an ssODN template is used, it is possible to design the template to bind to either the targeting strand or the nontargeting strand. The nontargeting strand is the strand that contains the protospacer adjacent motif (PAM), which is typically a sequence of 2–6 base pairs immediately following the DNA sequence targeted by the Cas9 nuclease. Although outcomes vary for different PAM sites, our research shows that HDR rates are often higher when the homology arms are identical to the nontargeting strand (Figure 2C).
Selecting a CRISPR Guide RNA
The first step in creating the perfect guide for insertion is selecting a suitable PAM site. We found that different PAM sites can have large variations in HDR rate (Figure 3). It is therefore recommended to measure HDR efficiency at a few different PAM sites, since the site closest to the insertion region is not necessarily the best for HDR.
However, the insertion cannot be too far away from the cut site. To test the relationship between the double-stranded break site and the location of the insertion, we inserted fragments at locations between 0 and 27 base pairs away from the cut site and plotted distance against the rate of HDR (Figure 3). The results show a sharp decrease in HDR rate as the distance from the cut site increases. This means that a balance between the efficiency of the PAM site and its proximity to the insertion will need to be determined empirically.
When studying HDR rates, it is also important to be aware of the effect of cell type on HDR. We found that the immortal cell line HEK-293 is a suitable cell type for HDR-mediated insertion. Conversely, several transformed cells and cancer cells show much lower rates of HDR. Primary cells can perform well in HDR studies, but results are likely to be variable.
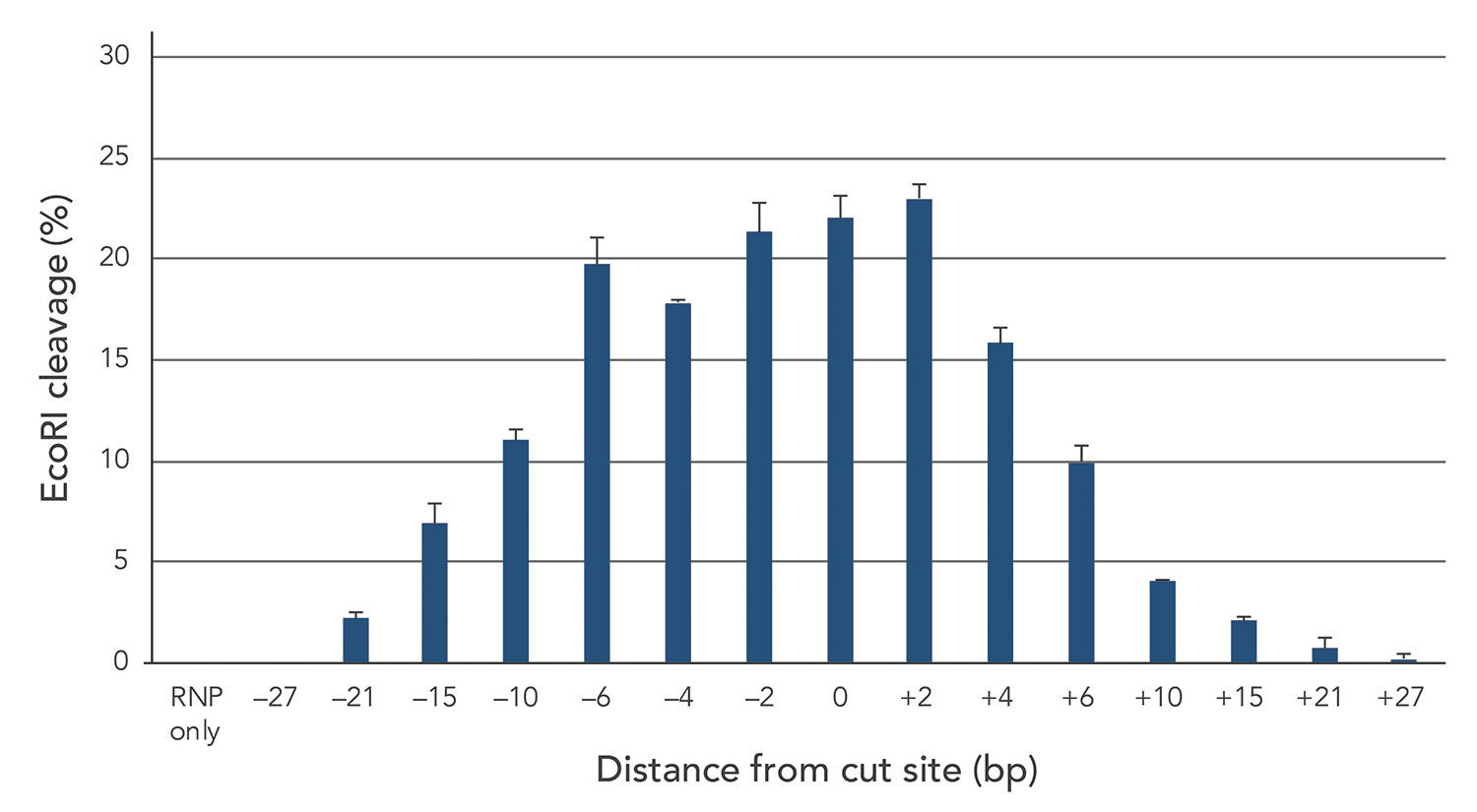
Figure 3. The distance between the double-stranded break and the location of the PAM site is a key parameter for HDR efficiency. Although HDR efficiency tends to decrease as this distance increases, the PAM site nearest the insertion region does not necessarily deliver the highest rate of HDR.
Error-Free Editing
Inserting exogenous sequences into the DNA of living cells is a key strength of CRISPR-Cas9. When reliable insertions with low error rates are required, HDR offers clear benefits over blunt-end incorporation by NHEJ. The results of our HDR optimization studies offers good insight into the parameters that determine how a template is inserted.
When it comes to optimization of the CRISPR guide RNA, finding the right PAM site is critical. Both the efficiency of the PAM itself and its proximity to the insertion site determine the rate of HDR incorporation—although some preliminary testing will be required. For the DNA template, Ultramer ssODNs are well suited for use in HDR experiments, as they offer higher efficiency, avoid blunt-end incorporation, and minimize cytotoxicity. Homology arms should be between 30 and 60 nucleotides long and may work slightly better when they match the nontargeting strand. These recommendations can help to improve CRISPR knock-in studies by enabling reliable, error-free editing.
Brian Wang, Ph.D. ([email protected]), is a genomic tools market development manager at Integrated DNA Technologies.