March 15, 2014 (Vol. 34, No. 6)
Exosomes no longer are being ignored. Identified over 30 years ago, these membrane-bound microvesicles have spent many years in the backwater of cell biology, regarded as only the cellular containers for disposing waste.
Shed by both normal and diseased cells, exosomes are now in the scientific spotlight. They have become the subject of numerous studies and the focus of two new scientific groups (American Society for Exosomes and Microvesicles and the International Society for Extracellular Vesicles).
To learn about current applications and future trends in exosomal studies, GEN reporter Cathy Yarbrough recently interviewed several scientists who specialize in exosome research. Those scientists are Suraj Bhat, Ph.D., Professor, Geffen School of Medicine, UCLA, Flavia Pichiorri, Ph.D., Assistant Professor, Ohio State University, Masato Mitsuhashi, M.D., Ph.D., CSO, Hitachi Chemical Research Center and Johan Skog, Ph.D., CSO, Exosome Diagnostics.
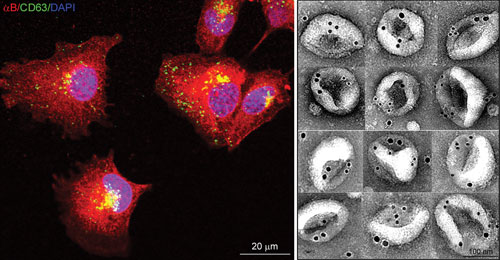
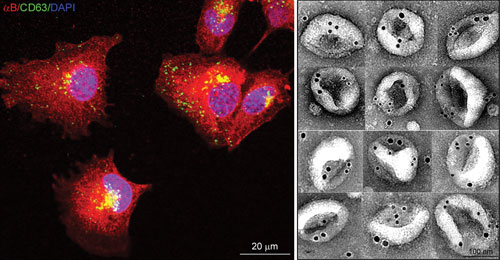
Left: Confocal Z-images of adult human retinal pigment epithelial (ARPE) cells expressing αB-crystallin (red) and CD63 (green), an endosome/exosome marker. Note co-localization in the perinuclear region. Right: Montage of electron micrographs showing immunogold-labeled (12 nm, αB; 18 nm, HSP70) native exosomes isolated from the culture medium of ARPE cells. Courtesy Rajendra K. Gangalum, Ph.D., UCLA.
GEN: Why did the scientific community not pay more attention to exosomes when they were identified 34 years ago?
Dr. Bhat
Exosomes initially may not have gained traction because cells pinch off a lot of membrane. So these microvesicles did not seem that novel. The scientific community paid attention when it became clear that there was a physiological basis to the production of exosomes. Most important, the technology to determine exosomes’ contents was developed.
Dr. Pichiorri
Cancer is a big driving force influencing the direction of research. So, when studies revealed that cancer cells secrete these easily accessible biomarkers, scientists were interested.
GEN: Why do you study exosomes?
Dr. Bhat
My lab’s primary research focus is alpha B-crystallin (alpha B), a small heat-shock protein that has been linked to Alzheimer’s disease, age-related macular degeneration, and several other neurodegenerative pathologies. It also has been shown to activate T cells in multiple sclerosis and inhibit platelet aggregation.
We did not expect alpha B to be secreted out of the cell. Determining that human adult retinal pigment epithelial cells (RPE) independent of the endoplasmic reticulum-Golgi protein export pathway secrete alpha B via exosomes was highly significant because it may explain many observations including the role of alpha B in immune function regulation and neural degenerations.
Since discovering the extralenticular expression of alpha B, we have been working to understand the physiological function of this protein. My lab has been trying to pinpoint the signals that trigger RPE cells to produce exosomes differentially from the apical and the basal side.
Because RPE works as a blood-retina barrier, we interrogate the instructions and the ensuing processes that assemble exosomes, two fundamental aspects of exosomal biogenesis that remain to be understood.
We think alpha B is essential for exosome biogenesis in human RPE cells in culture. Whether all cell-specific cargo, not just alpha B, has a “say” in the biogenesis of the exosomes that deliver them to their extracellular destinations is not yet known. Certainly, our data brings up a whole new way of looking at the endolysosomal pathway and the biogenesis of the endosomes and multivesicular bodies.
Dr. Mitsuhashi
Exosomes represent a new frontier in diagnostics with huge potential. In my lab, we are developing blood and urine diagnostics by quantifying mRNAs on exosomes.

Suraj Bhat, Ph.D. Professor, Geffen School of Medicine, UCLA
Dr. Pichiorri
Circulating miRNAs encapsulated in exosomes may serve as accurate prognosticators for multiple myeloma (MM), which is clinically very heterogeneous despite the homogenous morphologic appearance of the malignant plasma cells.
The research evidence for miRNAs’ role in MM is substantial. For example, studies have found that deregulated miRNAs target the coding genes implicated in MM, and that a global increase in miRNA expression is associated with poor prognosis.
My lab has determined that miRNAs levels likely are involved in multiple myeloma pathogenesis. This research was published in the Proceedings of the National Academy of Sciences in 2008. We also found the first evidence that miRNAs’ deregulation could be associated with mRNAs’ role in multiple myeloma.
By profiling 654 different miRNAs from multiple myeloma patients, we identified an association between three miRNAs (miR-16, miR-19b and miR-25) and multiple myeloma patients’ overall survival. At Ohio State University Medical Center, these miRNAs have been incorporated into the International Staging System, one of the methods now used to predict an multiple myeloma patient’s prognosis. Results thus far indicate that the miRNA-integrated score is highly predictive of survival duration.
In a manuscript now under review, we wrote that exosomes encapsulating the miRNAs associated with MM prognostication are generated not just by MM cells but also by peripheral blood cells such as monocytes, likely as a result of the body’s systemic reaction to the cancer.

Flavia Pichiorri, Ph.D. Assistant Professor, Ohio State University
Dr. Skog
I stumbled onto exosomes while studying cancer stem cells at Massachusetts General Hospital. I saw that these stem cells under the microscope had vesicles on them. They were exosomes. I wanted to know more.
We found both miRNA and mRNA in these vesicles and were surprised they were not described in the literature. Soon thereafter, a publication came out from the University of Gothenburg showing that exosomes from immune cells contained RNA. Spurred on by this finding, we continued to characterize the miRNA and mRNA in these vesicles and showed that the RNA could be used as a powerful biomarker even in serum that had been stored in biobanks for years. This led to the glioblastoma study, which we published in Nature Cell Biology in 2008.
Our understanding of exosome biology and exosomes’ clinical potential has advanced by leaps and bounds. At Exosome Diagnostics, we’ve developed technologies to analyze exosomes’ RNA and DNA content in biofluids from a range of different diseases, including prostate cancer, brain cancer, and lung cancer.
For example, if the genetic material in exosomes from a cancer patient contains an activating EGFR mutation, the oncologist could use this information to tailor the treatment with one of the EGFR-inhibiting drugs.

Johan Skog, Ph.D. CSO, Exosome Diagnostics
GEN: What are some of the milestone advances in exosome research?
Dr. Bhat
The discovery that exosomes can transform cells via lateral gene transfer has important implications for cell biology and suggests that these microvesicles may be used as vectors to transport molecular cargo from one cell to another. In fact, they could be employed as ligands for activating or deactivating target receptors, actions that could modulate gene activity.
Sharing molecular information between cells may be what keeps a multicellular organism together, and cells committing to disparate expression profiles and physiologies may be critical for sustaining different phenotypes including survival.
Dr. Mitsuhashi
For diagnostics, the milestone advances are the discovery of nucleic acids as well as proteins in the cargo transported by exosomes and the research findings showing that exosomes contain poly(A)+mRNA, which is protected from ribonucleases.
GEN: How do you envision the use of exosomes in diagnosis and therapy?
Dr. Skog
I hope that tumor-derived exosomes extracted from patients’ biofluids will be routinely used in blood or urine-based diagnostic tests, which have obvious advantages, particularly for tumors that now are diagnosed through surgical biopsies that are hard to come by.
In addition, because biopsies are so invasive, they are not suited to be used over time to monitor the tumors’ responses to specific therapies. Tumor dynamics change over time, so a biopsy conducted two years ago does not reflect the genetic profile of the patient’s cancer today.
Exosomes also have a potential application for delivering either genes or drugs to specific cells and are particularly useful for encapsulating drugs that are unstable.
Dr. Mitsuhashi
I strongly believe that within 10 years exosomes will be used for a wide variety of diagnostics not only in cancer, but also in cardiovascular, blood, brain, and kidney diseases; metabolic syndrome; etc. In fact, we are now capable of quantifying tissue/disease-specific mRNAs in plasma and urine. Before and after treatment, those values are widely different between control samples and samples from patients with these diseases.
In a paper just accepted by the Clinical Chemistry journal, we described the quantification of hematopoietic precursor cell-specific mRNAs in peripheral blood plasma. Testing that accurately quantifies these specific mRNAs may replace or reduce the frequency of brutal bone marrow aspiration.
In another project with control and patient urine samples, we successfully quantified exosomes that are specific to the glomerulus, proximal tubules, and distal tubules and ducts. Such testing will replace or reduce the frequency of invasive kidney biopsy.
Dr. Bhat
Understanding cell-type-specific control of exosome biogenesis may allow us to engineer exosomal content biologically. In less than 10 years, exosomes may be used as specific ligands or vectors to activate or modulate immune and stress responses, normal growth, and oncogenic pathways.
Dr. Pichiorri
A urine test for miRNA biomarkers will be developed to reliably distinguish cancer from normal cells. If cancer is indicated, the test will identify the specific form of malignancy.

Masato Mitsuhashi, M.D., Ph.D. CSO, Hitachi Chemical Research Center
GEN: What are the pressing exosome unknowns?
Dr. Bhat
What are the critical steps in the production of exosomes? What is the function of exosomes’ cargo? What is their relationship to disease phenotypes? How far does the exosome travel to deliver its cell-specific cargo to extracellular destinations? How much of the signaling and communication in the cells is achieved through exosomes? Does an interface between the endocrine system and exosomes exist, and if so, what is it?
Dr. Mitsuhashi
Standardization of exosome data and clinical correlation of the data are required.
Dr. Pichiorri
The phase markers on exosomes that can be used to distinguish these vesicles in a patient’s blood, urine, etc. as possibly associated with cancer or no disease must be identified.
Dr. Skog
For diagnostics, the isolation of quality exosomes has been a hurdle. At Exosome Diagnostics, we’ve spent many years of optimization to get to the point where we now can extract exosomes in a clinically useful way.
4th Annual Exosomes & Single Cell Analysis Summit
Johan Skog, one of the panelists in this roundup, is a keynote speaker in SelectBio’s upcoming Exosomes and Single Cell Analysis Summit to be held in San Diego on September 18 and 19. The meeting will present the very latest developments in the study of extracellular vesicles as well as the analysis of cells at the single-cell level.
It has become increasingly apparent that the differences between cells in the same population can be significant, and there is therefore a need for improved methods to capture and analyze individual cells, such as circulating tumor cells and cancer stem cells, and to study single cells for companion diagnostics development. This, coupled with new approaches for capturing exosomes and other classes of extracellular vesicles, is driving the field forward vis-à-vis development of biomarker assays, diagnostics development, as well as basic research on isolated individual components.