April 1, 2017 (Vol. 37, No. 7)
Lisa Heiden Ph.D. Director of Business Development MyBioSource
Think of Cancer as Criminal Activity, and You’ll Understand the Need to Properly Secure Evidence from Liquid Biopsies
Although circulating tumor cells (CTCs) and cell-free DNA (cfDNA) may be “bagged” in liquid biopsy samples, they may not stay intact through all the phases of an analytical workflow. If these biomarkers suffer degradation—during storage, transportation, nucleic acid extraction, or other sample preparation steps—they may skew assays toward erroneous or equivocal results. Plainly, this is no way to investigate a patient’s cancer, make a case for an individualized therapy, or monitor attempts at correction—to say nothing of guarding against treatment resistance or cancer recurrence.
The care and handling of liquid biopsy samples is something of a chain-of-custody problem. Fortunately, various links in this chain received special attention at the International Molecular Med Tri-Con event, which was recently held in San Francisco. This Cambridge Health Institute event was not solely devoted to liquid biopsies, but it did include a channel (“Circulating Tumor Cells and Liquid Biopsy”) and a symposium (“Circulating Cell-Free DNA”) of direct relevance to our subject.
Several of the event’s presentations are summarized below. All were selected to highlight key points in the analytical workflow, from the preanalytical “securing evidence” stage to the cancer management “post-judgment modification” stage. Preanalytical processing is vital for obtaining reliable downstream test results. CTC and cfDNA assays require that liquid biopsy samples “withstand stressors of preanalytical variables introduced in clinical settings,” asserted Landon Olp, Ph.D., R&D scientist, Streck.
“Isolated CTCs can be a better sample than a tumor biopsy,” insisted Steve Crouse, chief commercial officer, Vortex Biosciences. CTCs sloughed off from multiple locations during cancer progression represent multiple cell populations from both primary and metastatic tumors, capturing cancer heterogeneity.
Tumor cells release cfDNA into the bloodstream during cell death. “But cfDNA does not exist as freely dissociated DNA,” warned Hamid Khoja, Ph.D., principal scientist, Covaris. For example, cfDNA may be bound around nucleosomes and chaperone proteins, and it must be carefully extracted for downstream analysis.
Analysis technologies such as next-generation sequencing are not without error. “The need for sensitivity and accuracy is critical,” advised Dale Yuzuki, market development director, SeraCare Life Sciences. “Without standards, people won’t know if a test and claimed sensitivity [are] real or not.”
Phillip G. Febbo, M.D., CMO, Genomic Health, added that “you have to understand the performance of your assay with respect to clinical samples and the likely allele frequencies you might see in those clinical samples.”
Stabilizing cfDNAs in Sample Tubes
“There are many preanalytical variables, but sample storage and transportation probably have the greatest impact on cfDNA assays,” remarked Streck’s Dr. Olp.
Most doctors’ offices can’t perform cfDNA tests in house. Blood samples are collected, shipped to a central testing lab, and often batched for cost savings. “It could take a week or longer from the time a blood sample is drawn until tested using a cfDNA assay,” lamented Dr. Olp.
Shipping and storage places a high level of stress on the sample. Blood cells lyse in a standard EDTA blood collection tube; releasing cellular DNA into the plasma, the cfDNA space.
“The already rare mutant cfDNA, your biomarker, becomes even more diluted,” informed Dr. Olp. “In essence, your needle in a haystack becomes a needle in a field of haystacks, and the assay results can be unreliable due to the instability of the sample.”
To resolve this problem, Streck provides a specialized blood collection tube (BCT), the Cell-Free DNA BCT®. It contains a reagent that can stabilize nucleated cells right in the tube, enabling the sample to withstand the stressors of the preanalytical variables for up to 14 days at room temperature. The release of cellular DNA from white blood cells breaking up is prevented, and fragile CTCs don’t degrade.
“Stabilization allows Cell-Free DNA BCT’s to be applicable for both cfDNA and CTC biomarkers,” emphasized Dr. Olp. “The original proportions of cfDNA and CTCs present in the sample will remain consistent for up to two weeks, giving you time before you begin processing that sample in a liquid biopsy assay.”
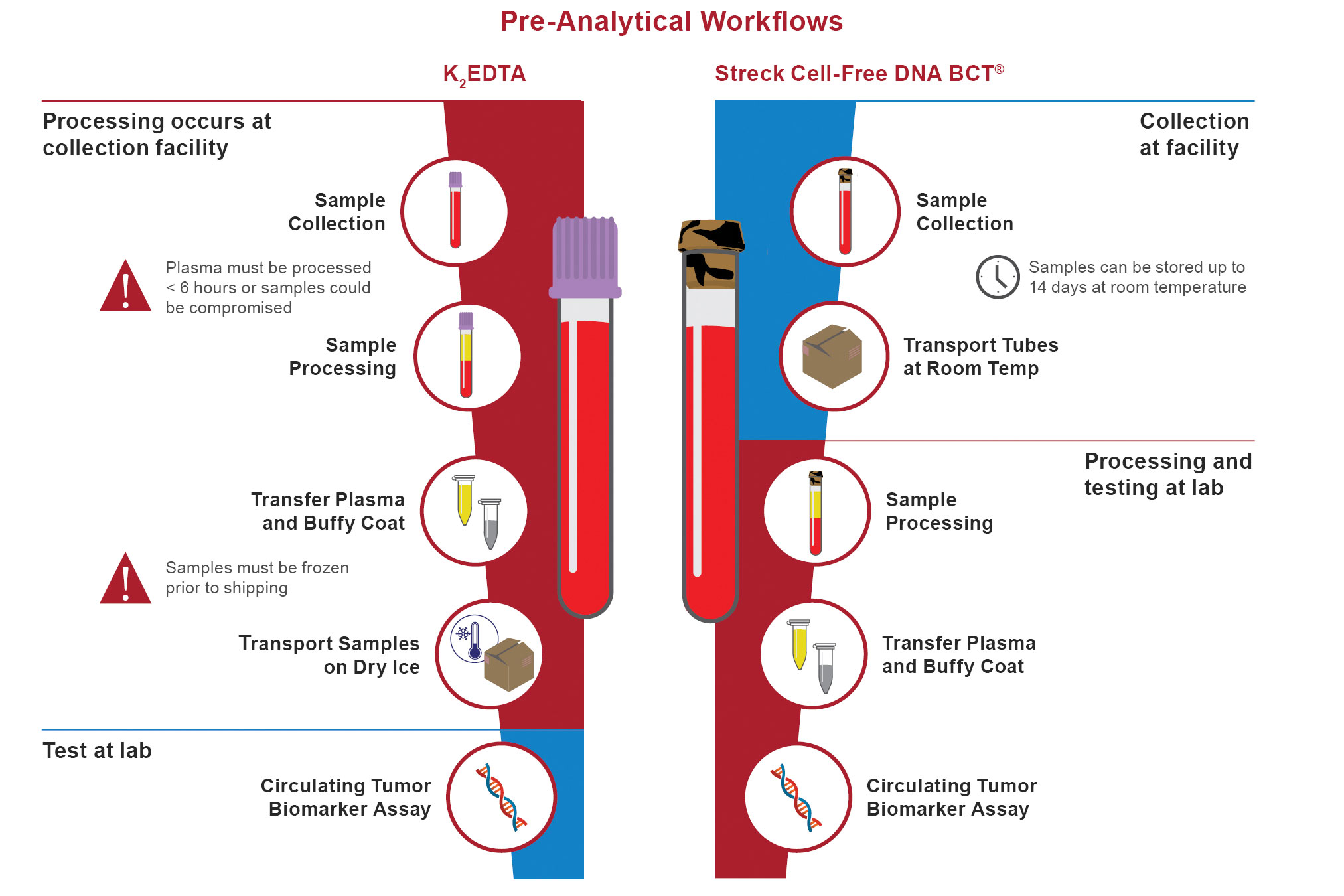
Streck’s Cell-Free DNA BCT simplifies collection, storage, and shipping of whole blood samples for circulating tumor biomarker assays. The stabilizing reagent included in the Cell-Free DNA BCT enables nucleated blood cells to withstand the stressors of preanalytical sample handling up to 14 days postphlebotomy—protecting the original proportions of cell-free DNA and circulating tumor cells in the sample.
Trapping CTCs in Microvortices
“The automated VTX-1 Liquid Biopsy System from Vortex Biosciences evaluates both the physical properties of cells and the characteristics of molecular markers,” noted Crouse. Many CTC isolation technologies bind EpCAM, an epithelial marker, to capture CTCs. Yet, many cancers don’t express EpCAM or lose expression during metastasis. Other technologies use a filter, which only targets the larger CTCs and can get clogged.
The VTX-1 utilizes microfluidics to efficiently isolate CTCs directly from whole blood. “The VTX-1 flows blood down a microchannel which widens along a portion of its length, creating a reservoir. Between the narrowings at the inlet and outlet, microvortices form, similar to the vortices that may form in a river and trap leaves in looping paths,” explained Crouse. The VTX-1 vortices trap the larger and more deformable cells such as CTCs, stably holding them in a swirling current while the rest of the blood cells flow past.
“Collecting based on physical properties captures a diverse population of cancer cells and, we believe, a more clinically relevant sample than affinity-based approaches,” remarked Crouse. Unlike affinity-based or filter-based collections, cells are not stuck onto anything like channels, membranes, or magnetic beads.
Since the VTX-1-captured cells stay in suspension and experience pressures similar to those they would encounter in circulation, they remain intact and similar to how they were in the bloodstream. CTCs are released from the Vortex microfluidic chip and are immediately available for genomic, RNA, protein, or live-cell assays, allowing for a holistic picture of the cancer.
The VTX-1, which accommodates 0.2–16 mL samples, has several exciting capabilities. For example, it can capture CTCs from the tiny liquid biopsy samples of patient-derived xenograft (PDX) mouse models, “where the VTX-1 offers an opportunity to get access to cancer biology in a way that wasn’t available before,” exclaimed Crouse.
The VTX-1 enables synergistic CTC and cfDNA approaches. “A Streck Cell-Free DNA BCT can be used to ensure cfDNA remains stable. After pulling off the plasma (containing cfDNA), you can process the remaining blood through the VTX-1 and collect the CTCs,” explained Crouse. This enables mutation profiling of both the CTCs and cfDNA from a blood tube, potentially providing a more informed understanding of the patient’s cancer.
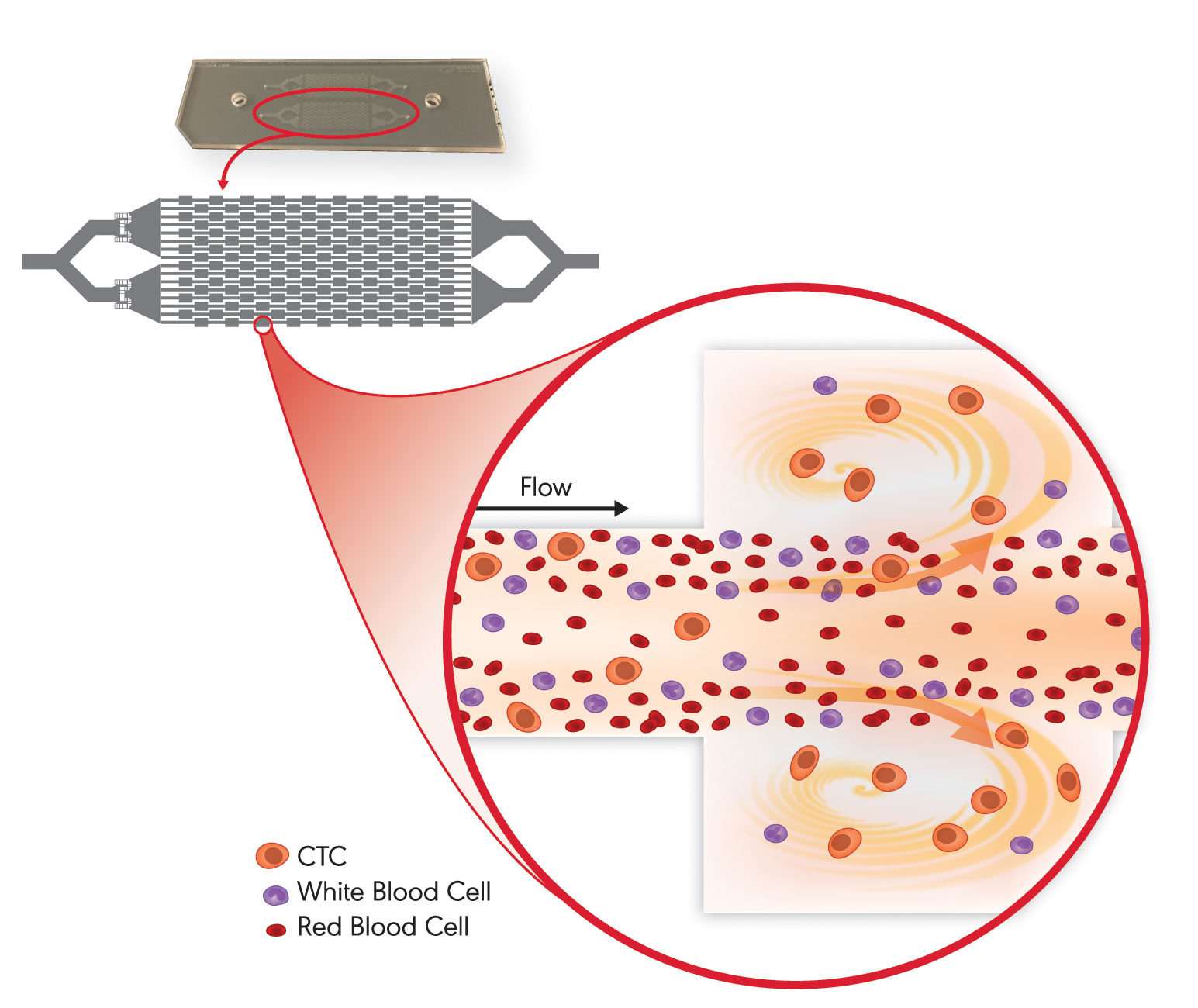
With Vortex Biosciences’ VTX-1 Liquid Biopsy System, a blood sample flows down microchannels in a microfluidic chip, which contains chambers that sustain microvortices. These microvortices trap and retain the larger and more deformable circulating tumor cells (CTCs) while allowing blood cells to flow past. Once isolated, the CTCs can be released for downstream analysis.
Soundly Standardizing Extraction
Clinical decisions are made based on quantitative cfDNA results. “Although analytical technologies become more sensitive every year, the sample preparation workflows feeding these instruments are often low tech, antiquated, and inefficient,” cautioned Covaris’ Dr. Khoja.
The Covaris Adaptive Focused Acoustics™ (AFA) platform normalizes diverse sample specimens and standardizes preanalytical sample preparation. AFA uses focused acoustic energy to actively control cavitation. Cavitation is caused by the conversion of acoustical to highly tuned mechanical, sheer force energy. “That’s how you get a variety of beneficial effects, such as DNA shearing and extraction, cell lysis, and solvation of a boundary layer,” explained Dr. Khoja.
AFA minimizes chemical variability introduced during cfDNA extraction and purification, while maximizing cfDNA yields from precious clinical samples. These features enable the power of sensitive analytical instruments to be realized so that these devices may facilitate the development of more sensitive and relevant next-generation clinical applications.
AFA is highly controllable. The non-contact isothermal AFA technology is very tunable, such that DNA can be randomly sheered to many different lengths (100 bp–5 kb, depending on the library preparation needs). “The uniformity of coverage obtained with an AFA-prepped sample enables lower sequencing depth, speeding sequencing and reducting its cost,” remarked Dr. Khoja.
The cfDNA concentration is highly variable between patients, tumor types, stages, and treatment conditions. Standardization of all preanalytical protocols, including blood collection, is critical for optimal analytical results. “We recommend Streck Cell-Free DNA BCT because it brings a level of control over the collection, stabilization, and shipping of precious blood samples,” stated Dr. Khoja.
Covaris develops kits for standardizing cfDNA extraction. The truXTRAC cfDNA™ kits are designed for actively dissociating and extracting cfDNA from histone-cfDNA and other protein-cfDNA complexes occurring in cfDNA BCT-stabilized plasma, and they enable scalable and automatic high-throughput sample processing.
“DNA sequencing confirmed that truXTRAC cfDNA-processed samples had higher library complexity, more thoroughly mapped reads, greater coverage uniformity, and better variant sensitivity than cfDNA samples obtained from passive extraction methods,” concluded Dr. Khoja.
Getting to the Ground Truth with Reference Standards
“Mutant DNA may be less than 0.5% of total cfDNA shed into the bloodstream, and tests will need to get sensitivity down to about 0.1%,” said SeaCare Life Sciences’ Yuzuki, who added that the need for sensitive tests is imperative. He questioned, however, whether many investigators were capable of detecting one mutant molecule in ~1,000 wild-type molecules. “If your technology is throwing off false negatives or positives due to factors beyond your control, it can dirty up the clinical trial results,” he warned.
SeraCare engineers biosynthetic cfDNA reference standards. The multiplexed Seraseq™ Circulating Tumor DNA Mutation Mix kits contain nine actionable mutations. The standards assist in liquid biopsy assay development, validation, and monitoring, enabling researchers to know the ground truth. “If your standard is sensitive down to 0.1%, then a mutation at 0.1% in your sample can reliably be picked up,” asserted Yuzuki.
Knowing the limit of detection of your cfDNA assay is an important ground truth, too. “Perhaps your assay is reliably down to 0.6%. Then over time, variables change, such as reagents, operators, and next-generation sequencing bioinformatics pipelines,” commented Yuzuki. But a 0.6% reference standard won’t change, and so it can be used as a tool to revalidate your assay.
The standards are engineered by blending actionable biosynthetic DNA targets with a single, well-characterized wild-type genomic background derived from the GM24385 cell line, which was contributed by a volunteer in the Personal Genome Project. Allelic frequency is strictly controlled. The standards are commutable to native samples and behave similarly to native plasma mutant cfDNA in library preparations.
Liquid and solid tumor biopsies may be used as complementary approaches during the course of cancer management. The clinically important, actionable mutations in SeraCare’s solid mutation mixes include 25 single-nucleotide variations and 13 insertion/deletion polymorphisms (INDELs), such as a 15-bp EGFR deletion and a 12-bp ERBB2 insertion, and two structural variants. The mixes of 40 mutations are offered in three allelic ratios (10%, 7%, and 4%) in both high and low concentrations.
“Typically, assays will reach a 5% allele frequency sensitivity. Now, laboratories can validate their assays slightly above and slightly below a claimed limit of detection using 4% and 7% Seraseq Mutation Mix kits, respectively,” concluded Yuzuki.
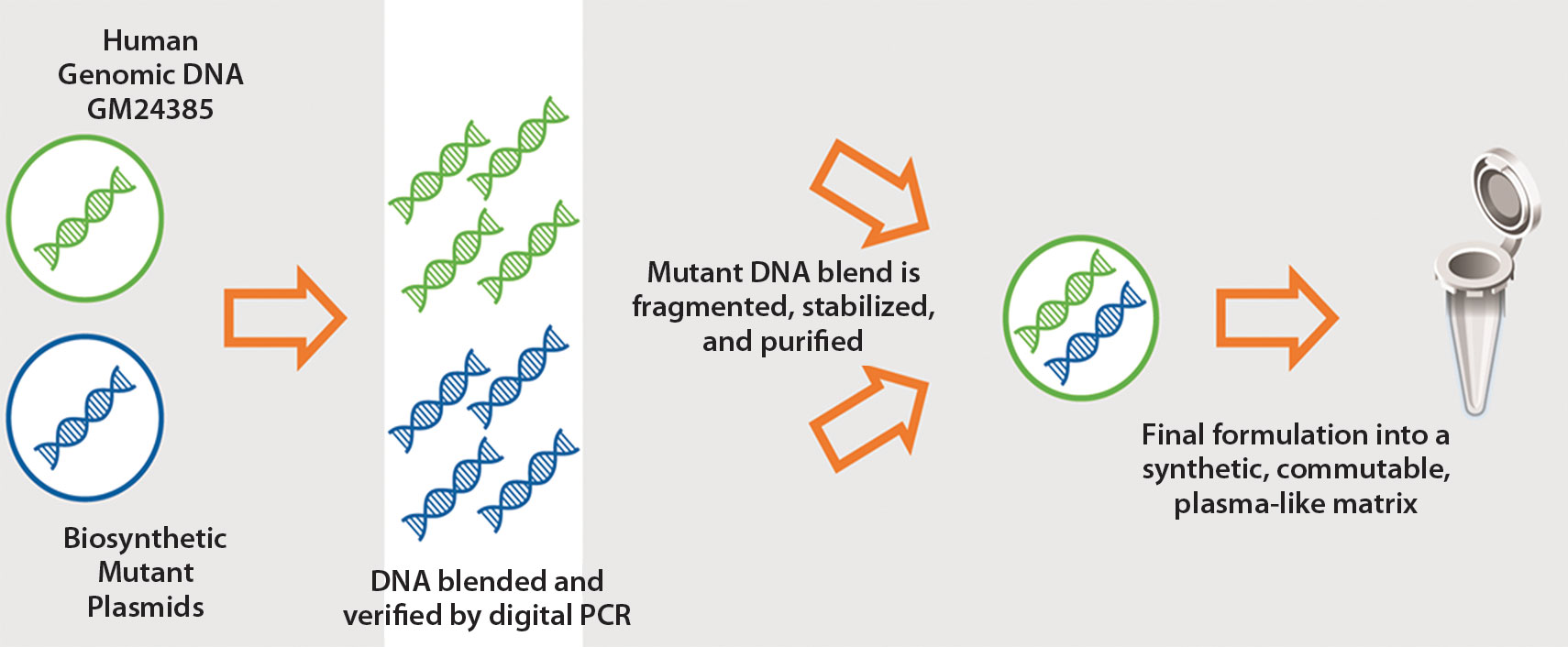
SeraCare Life Sciences manufactures patient-like mutant cell-free DNA (cfDNA) reference material. The company indicates that its Seraseq Circulating Tumor DNA-I Reference Material is composed of a set of nine actionable “driver” oncology mutations that are blended at a precise allele-frequency ratio to GM24385 human reference DNA, fragmented to approximately 170 base pairs, stabilized via a patent-pending method, and mixed into a human plasma-like matrix.
Utilizing cfDNA in Cancer Management:
The benefits of a growing number of cancer therapies depend on the presence of specific variants. It’s essential to understand which variants are present in patient tumors and “to understand that with confidence,” emphasized Genomic Health’s Dr. Febbo.
“The cfDNA space overcomes significant challenges of tissue accessibility,” he continued. About one-third of lung cancer patients only have enough tissue for up to two molecular tests, or their tissue is insufficient for molecular testing. Accessing tissue and repeated procedures in patients with comorbidities can be challenging. Detecting variants through cfDNA can bring precision medicine to patients who otherwise wouldn’t be able to benefit.
There are at least seven known genes in lung cancer with variants. “If those variants are present, we know those patients will benefit from specific therapies targeting their variant,” stressed Dr. Febbo. “These patients might also be able to avoid the overuse of chemotherapy.” He indicated that lung cancer is probably the lead clinical indicator, where Genomic Health and others are focusing on bringing products into the clinical realm.
The performance of liquid biopsies comes down to understanding clinical specificity and sensitivity. “Patients and physicians should have confidence that the variants we call are present and not false positives,” explained Dr. Febbo. “We make sure patient-level specificity is greater than 99%.”
Patients also want to have confidence in sensitivity, the level at which a variant can be detected if the variant is present. The signal from the tumor can be quite quiet, as mutant tumor DNA is often below 1% or even 0.5% of total cfDNA.
An understanding of the technology underlying your cfDNA test is essential. “The strengths and limitations of the technology are an incredibly important part of the test,” insisted Dr. Febbo.
Genomic Health has qualified its Oncotype SEQ® Liquid Select clinically actionable 17-gene tumor mutation panel against each of the common types of variants: single-nucleotide variations, INDELs, copy number variations, and rearrangements. Next-generation sequencing is used for a “comprehensive and robust assessment of each gene,” Dr. Febbo concluded.
Ensuring Inter-lab Reproducibility in Quantification of ctDNA
Reproducibly quantifying DNA copies between different laboratories is inherently challenging, but a team at LGC, a U.K.-based measurement institute, reports that digital PCR may be able to clear this longstanding hurdle.
Of the 21 international labs that participated in a recent study, all yielded the same results. The absolute quantities of a KRAS point mutation were accurately measured at all fractional abundances (ranging from 12–0.2% mutant), all without calibration.
It is significant that digital PCR can be used to accurately and consistently measure such rare mutant sequences, especially considering that this is not achievable with quantitative PCR, a popular method that is difficult to perform reproducibly due to challenges with calibration. Jim Huggett, Ph.D., principal scientist at LGC, senior lecturer at the University of Surrey and managing editor of the Journal of Biomolecular Detection and Quantification, coordinated the study with Alexandra Whale, Ph.D., a senior researcher at LGC.
Dr. Huggett’s team evaluates the measurement of molecular biomarkers in clinical research and diagnostics. According to Dr. Huggett, the KRAS point mutation was chosen as the test case precisely because it is difficult to measure and a clinically relevant ctDNA target. His group published the study in the journal Analytical Chemistry.
All 21 participating laboratories used a Droplet Digital PCR platform and wet-lab validated rare mutation detection assays from Bio-Rad Laboratories to quantify DNA in blinded samples. Droplet Digital PCR is performed by initially partitioning the DNA into thousands of droplets using water and oil.
Absolute quantification is performed by counting the positive, DNA containing, partitions after the PCR has been performed. Study participants included both experienced digital PCR users and researchers who were newer to the technique.
Dr. Huggett’s team prepared and characterized all the samples, including positive and negative controls, and then sent them to the international sites for blinded testing. The tests were conducted in hospital, research and industry laboratories.







