Miniaturization and automation are bringing new tools and strategies to cell-culture process development for production of therapeutic proteins. In addition to automating existing culture platforms such as flasks and shaken well plates, novel high-throughput predictive scale-down platforms are also being developed.
Automating such processes can increase experimental throughput and accelerate process development to drive therapeutic proteins to clinical trials faster. Having such platforms allows for new methodologies in process development.
Traditional cell culture process development (PD) uses flasks and bench-scale bioreactors to determine optimal media formulations and process parameters. Because these tools are not amenable to high-throughput experimentation, the traditional PD approach is to carefully design limited experiments in order to manage resources, time, and budget.
These limited experiments are usually designed to collect as much information as possible. Typically, experiments are designed based on experience, cell-culture knowledge, and statistics, while information such as viable cell density, metabolites, and protein quantity and quality are collected.
Here the paradigm is low experiment throughput with high data content; find out as much as possible with the limited number of experiments to determine optimal process conditions. Yet this approach may not identify critical interactions, say between a key media component and pH, or even identify the true optimum in the operating space. Further, extensively characterizing local process maxima is unlikely to give hints to the existence of a global maximum.
High-throughput Strategy
With a high-throughput cell culture platform, larger statistically designed studies can be conducted. With the ability to run hundreds of experiments in parallel, clones can be screened against various media formulations and/or process variables, effects of and interactions between multiple media components can be extensively examined (e.g., a full factorial or large fractional factorial), process operating space can be thoroughly mapped out to determine optimal conditions, and sensitivity analysis can be conducted on variables themselves.
In other words, a high-throughput platform allows users to take full advantage of statistically designed experiments to obtain extensive information toward process optimization and characterization. Statistically designed studies can provide large amounts of information without having to analyze in detail each cell culture experiment, thus avoiding the possible bottleneck of being overwhelmed in sample and data analysis with high-throughput experimentation.
So instead of performing small DOE (design of experiment) or one factor at a time studies and thoroughly characterizing a relatively small number of bioreactor runs, a better PD strategy for high-throughput cell culture tools is to perform large DOE studies to extensively map operating space without exhaustively analyzing each design point. Once optimal conditions have been identified, they can then be tested with a traditional system such as a bench-scale bioreactor, where further characterization can be performed.
What follows is a simple case study using this high-throughput paradigm. BioProcessors’ (www.bioprocessors.com) SimCell™ MicroBioreactor platform is used in a process-development exercise as a high-throughput cell culture tool after a process is scaled-down to establish predictability of the system. A simple study is conducted to optimize media, pH, and temperature profile, which is scaled up to a bench bioreactor where the improved process is characterized in depth.
Scalable High-throughput Cell Culture
SimCell MicroBioreactor technology is designed to simulate large bioreactor batch and fed-batch processes at submilliliter volumes. The ability to monitor and control parameters such as pH, temperature, and dissolved oxygen (DO) with SimCell improves scale-up predictability over conventional systems such as flasks and shaken well plates.

Six submilliliter bioreactors are contained in one MicroBioreactor Array (MBA; Figure 1), each housing noninvasive optical sensors for pH and DO and constructed with transparent, gas-permeable membranes for exchange of culture gases (e.g., O2, CO2). MBAs are manipulated with a high-capacity robotic system automating inoculation, incubation, sampling, feeding, and culture process monitoring and control.
To establish scalability of this platform, a process was scaled down from a bioreactor. For this case study, a CHO cell line producing human interferon-g (IFNg) was grown in a six-day batch process in two bench bioreactors and six SimCell MicroBioreactors. Growth, protein production, and metabolism were compared between the two systems.
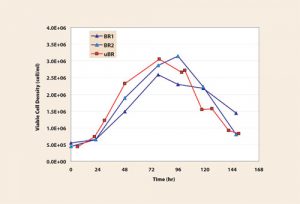
Cell growth (Figure 2), metabolism, and IFNg production results show good comparability between the two systems. Specifically, similar integral of viable cell concentration (IVCC) and maximum cell densities were achieved on day four. Further, IFNg yield and IFNg production rates were similar, and the ratio of moles lactate produced per mole glucose consumed was nearly 1 in all cases. Based on these results, the CHO IFNg process in MicroBioreactors is concluded to be a good model for the bench-scale bioreactor.
PD Optimization
With scalability established, an experiment was designed to determine optimal pH and blend of two commercially available serum-free media formulations, A and B. The software program Design Expert 6.0 (Stat-Ease) was used to create a factorial experiment and to analyze the results. Choosing maximum cell density as a metric will indicate which conditions yield favorable growth, whereas choosing final protein yield as the other metric will reflect productivity.
Further, results can be analyzed to identify conditions under the constraints of optimizing both metrics, in effect identifying conditions that result in high, specific production (protein produced per cell) of IFNg.
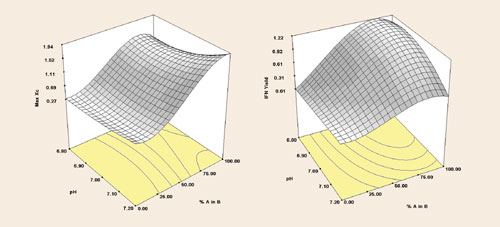
The experiment was run as a full factorial on a SimCell platform with six replicates per condition, for a total of 126 cultures. By choosing only two metrics, analysis was kept relatively simple to minimize the amount of data analysis and sample processing upon harvest. Nonetheless, the platform collected over 21,000 data points from this experiment. Response surfaces (Figure 3) identify optimal conditions to be 80% media A in B and pH controlled to 6.8 and a process that is more sensitive to blend than pH.
A second, smaller study examined the effect of temperature shift on IFNg yield. Simulating a batch process using SimCell MicroBioreactors with pH controlled to pH 6.8 and CHO cells grown in the optimal blend and shifting temperature from 37 to 34°C on day three indicated a potential twofold increase in protein yield.
Scaling Up and Process Characterization
Once optimal media, pH, and temperature conditions were identified, the next step was to run the new process in 3 L bioreactors to check scalability and to characterize the improved process. Two bioreactor process runs were performed: the first used the optimal media blend and pH, while the second was identical to the first but included the temperature shift.
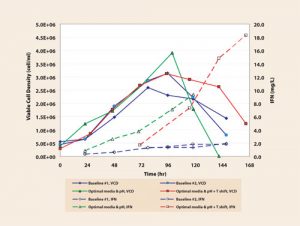
Growth curves and IFNg yield (Figure 4) from these two runs confirm improved performance predicted by the high-throughput MicroBioreactor experiments. Optimal media blend and pH increased maximum cell density up to 1 million cells/mL (a greater than 36% increase), while adding a temperature shift further extended culture viability up to two days and increased IVCC by 27%.
More dramatic results are seen with IFNg production as optimal blend and pH increased yield 4.8-fold, while the temperature shift further doubled yield to over 18 mg/L. Overall the yield increased by over ninefold, and specific productivity increased nearly sevenfold based on experiments conducted in less than four weeks.
Bench-scale bioreactors are a convenient tool to allow for in-depth analysis and to investigate effects of optimal conditions determined with scale-down models as there are fewer samples to analyze and the amount of data is easily manageable.
In addition to characterizing growth and protein production, metabolites and specific production rates were examined in detail on the bioreactor scale-up runs. Glucose was not fully consumed for baseline, with about 2 g/L remaining, yet under optimal conditions, glucose was not only fully consumed but the amount of lactate produced per glucose consumed decreased from about 1 to less than 0.8 mole/mole. Further, the ratio of IFNg produced per glutamine consumed increased significantly by 10-fold, from 2.8 to 28 mg/g.
Understanding dependence, sensitivity, and robustness of a cell culture bioprocess is critically important for technology transfer, production scale-up, and regulatory certification.
To be valuable, a scale-down model must be able to rapidly interrogate a wide process operating space. To improve predictability of the model, it must be able to control process variables such as temperature, pH, dissolved gases, and media composition.
With advances in miniaturization and automation in cell culture industry, PD scientists and engineers now have tools to rapidly and thoroughly optimize cell culture process space.
With statistically designed cell culture studies, extensive characterization of each design point is not necessary as process sensitivity to factors, interactions between factors, and optimal values can be determined by examining a few key metrics, thus avoiding analyzing large amounts of samples and data.
A new strategy can now be taken; one that uses high-throughput tools to rapidly screen process variables in conjunction with traditional, low-throughput tools for in-depth analysis and characterization.
H. Brett Schreyer ([email protected]) is principal engineer, Scott E. Miller is principal scientist, and Seth Rodgers is CTO at BioProcessors.






