Over the past two years, machine learning has revolutionized protein structure prediction. This has been led by experimentally characterized de novo protein designs that have been generated using physically based approaches. Now, a similar revolution in protein design is described.
This work is published in Science, in the paper, “Robust deep learning–based protein sequence design using ProteinMPNN.”
“Proteins are fundamental across biology, but we know that all the proteins found in every plant, animal, and microbe make up far less than one percent of what is possible. With these new software tools, researchers should be able to find solutions to long-standing challenges in medicine, energy, and technology,” said Baker.
Recently, powerful machine learning algorithms including AlphaFold and RoseTTAFold have been trained to predict the detailed shapes of natural proteins based solely on their amino acid sequences. To go beyond the proteins found in nature, Baker’s team members broke down the challenge of protein design into three parts and used new software solutions for each.
First, a new protein shape must be generated. In a paper published July 21 in the journal Science, the team showed that artificial intelligence can generate new protein shapes in two ways. The first, dubbed “hallucination,” is akin to DALL-E or other generative AI tools that produce output based on simple prompts. The second, dubbed “inpainting,” is analogous to the autocomplete feature found in modern search bars.

“Neural networks are easy to train if you have a ton of data, but with proteins, we don’t have as many examples as we would like. We had to go in and identify which features in these molecules are the most important. It was a bit of trial and error,” said Justas Dauparas, PhD, a postdoctoral fellow in the Baker lab at the Institute for Protein Design.
Third, the team used AlphaFold, a tool developed by Alphabet’s DeepMind, to independently assess whether the amino acid sequences they came up with were likely to fold into the intended shapes.
“Software for predicting protein structures is part of the solution but it cannot come up with anything new on its own,” explained Dauparas.
“ProteinMPNN is to protein design what AlphaFold was to protein structure prediction,” added Baker.
In another paper appearing in Science on Sept. 15, a team from the Baker lab confirmed that the combination of new machine learning tools could reliably generate new proteins that functioned in the laboratory.
“We found that proteins made using ProteinMPNN were much more likely to fold up as intended, and we could create very complex protein assemblies using these methods,” said Basile Wicky, PhD, a postdoctoral fellow in the Baker lab.
The authors wrote that, on native protein backbones, ProteinMPNN has a sequence recovery of 52.4%, compared to 32.9% for Rosetta. And, they say that “the amino acid sequence at different positions can be coupled between single or multiple chains, enabling application to a wide range of current protein design challenges.”
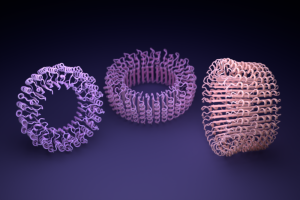
Among the new proteins made were nanoscale rings that the researchers believe could become parts for custom nanomachines. Electron microscopes were used to observe the rings, which have diameters roughly a billion times smaller than a poppy seed.
“This is the very beginning of machine learning in protein design,” said Baker. “In the coming months, we will be working to improve these tools to create even more dynamic and functional proteins.”







