April 1, 2006 (Vol. 26, No. 7)
RNA Amplification & cDNA Synthesis Enables Sensitive qRT-PCR of Low-Abundance Transcripts
Ideally, gene expression studies by quantitative RT-PCR (qRT-PCR) should be performed using RNA from homogeneous cell populations. Frequently, this means performing qRT-PCR using picogram amounts of total cellular RNA and, in some instances, using total RNA from a single cell.
Technologies such as laser capture microdissection and fluorescence-activated cell sorting make it possible to obtain small cell populations. However, qRT-PCR detection of even high-abundance transcripts from a minute number of cells is challenging, as evidenced by high cycle threshold (CT) values and a lack of consistency between replicates. Often low- and medium-abundance transcripts are not detected at all. Additionally, only a limited number of qRT-PCR reactions can be performed using the total RNA that can be isolated from a small sample.
Epicentre Biotechnologies (www.epibio.com) recently introduced the MessageBooster cDNA Synthesis Kit for qPCR, which enables sensitive and reproducible qRT-PCR of even low-abundance transcripts using total RNA from small cell populations (1 to 50 cells). A MessageBooster reaction amplifies the poly(A) RNA present in a total RNA preparation and then converts the amplified RNA to cDNA, which can be used without purification for qPCR. A MessageBooster reaction preserves the relative transcript abundance of the sample and yields enough cDNA to perform many more qPCR reactions than can be obtained from untreated total RNA samples.
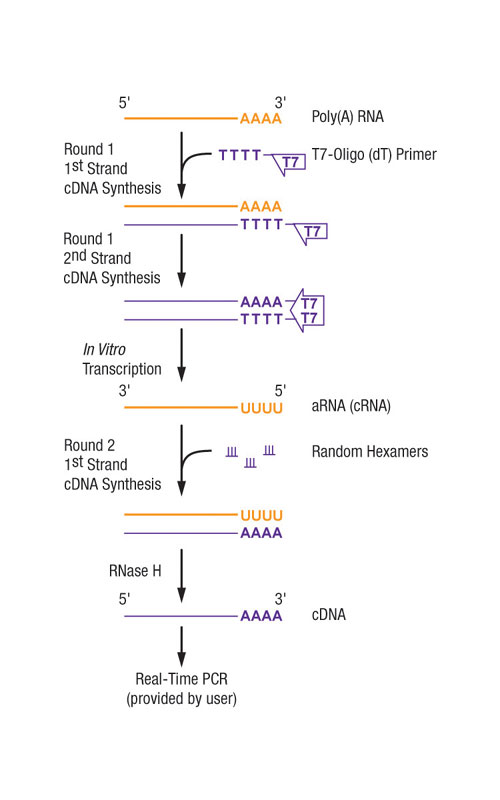
The MessageBooster cDNA Synthesis Kit for qPCR utilizes an improved linear poly(A) RNA amplification process (Figure 1) first described in the laboratory of James Eberwine. Briefly, the poly(A) RNA in a total RNA sample from as little as one cell is converted to double-stranded cDNA.
The cDNA is subsequently used as template in a high-yield T7 in vitro transcription reaction that produces antisense RNA (aRNA, also called cRNA). The aRNA is then reverse transcribed into single-stranded, sense cDNA that can be used without purification for qPCR. A MessageBooster reaction can be completed in one day.
Fig.1: A MessageBooster(tm) Kit reaction utilizes a linear RNA amplification process to amplify the poly(A) RNA(mRNA) from the total RNA of as little as 1 cell and then converts the aRNA produced to single-strand sense cDNA that is ready for qPCR.
Preservation of Transcript Abundance
Preservation of the relative transcript abundance following an RNA amplification process is critical to ensure accurate qRT-PCR results. Figure 2 shows the normalized difference in expression between Universal Reference RNA and adult skeletal muscle RNA (DDCT) for 20 transcripts tested before and after a MessageBooster reaction are plotted.
The normalized difference in expression was calculated as the difference in CT between test amplifications and normalizer amplifications (DCT). The high correlation coefficient, R2=0.997, demonstrates that the cDNA produced by a MessageBooster reaction maintains the relative transcript abundance of the sample.
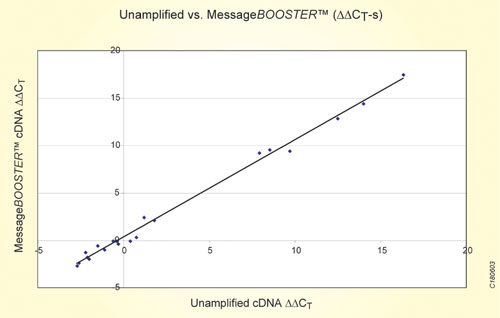
Fig.2: Scatter plot of the difference in relative transcript abundance of 20 mRNA targets in Human Reference RNA and in adult skeletal muscle RNA before and after a MessageBooster reaction.
Sensitive qPCR of Low-Abundance Transcripts from a Single Cell
The poly(A) RNA in 10 pg of total normal rat kidney (NRK) RNA (about the amount of total RNA in one cell) was amplified, and the aRNA produced was converted to cDNA in a standard MessageBooster reaction (MessageBooster cDNA).
In a parallel reaction, the poly(A) RNA in 10 pg of total NRK RNA was converted to cDNA without benefit of a MessageBooster reaction (unamplified cDNA). PCR primers and sequence-specific probes for three transcripts present in NRK cells, porphobilinogen deaminase (PBGD; a low-abundance transcript), pre-B-cell leukemia transcription factor 2 (PBX2; a low-to-medium-abundance transcript), and beta-2-microglobulin (B2M; a medium-to-high-abundance transcript) were synthesized and qPCR performed using MessageBooster cDNA and unamplified cDNA.
Figure 3 shows the quantification graph obtained for qPCR of the low-abundance PBGD transcript using MessageBooster cDNA and unamplified cDNA. MessageBooster cDNA yielded significantly improved detection sensitivity, as determined by a reduction in the CT value when compared to the qPCR result obtained using unamplified cDNA. Greatly improved detection of the PBX2 and B2M transcripts was also observed using MessageBooster cDNA compared to unamplified cDNA (data not shown). SYBR Green I dye detection chemistry provided comparable results to those using sequence-specific fluorescent probes.
The number of qPCR reactions that can be obtained using the cDNA produced by a MessageBooster reaction was found to be dependent on two factors: the amount of total RNA added to the MessageBooster reaction and the abundance of the transcript(s) of interest. Table 1 summarizes our estimate of the number of qPCR reactions that can be obtained using cDNA produced from a MessageBOOSTER reaction containing input total RNA from ~1, 10, and 50 NRK cells.
The qRT-PCR detection sensitivity of low-abundance transcripts (as well as medium- and high-abundance transcripts) from very small samples (1 to 50 cells) can be greatly increased using the MessageBooster cDNA Synthesis Kit for qPCR. A MessageBooster reaction preserves the relative transcript abundance of the sample and produces enough cDNA for up to hundreds of qPCR reactions.
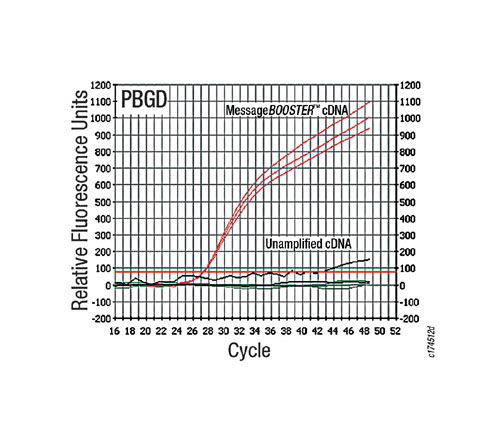
Fig.3: cDNA produced from 10pg of total RNA using eh MessageBooster(tm) cDNA Synthesis Kit for qPCR significantly improves the sensitivity of detection of a low-abundance transcript compared to cDNA produced from 10pg of unamplified RNA.