Researchers from the Moscow Institute of Physics and Technology (MIPT), the Skolkovo Institute of Science and Technology (Skoltech), and the University of Southern California (USC) say they have developed a new computational method for the design of thermally stable G protein-coupled receptors (GPCR) that are of great help in creating new drugs. The method has already proved useful in obtaining the structures of several principal human receptors, according to the team.
An overview (“Computational design for thermostabilization of GPCRs”) of the new method was published in Current Opinion on Structural Biology.
“GPCR superfamily is the largest clinically relevant family of targets in the human genome; however, low thermostability and high conformational plasticity of these integral membrane proteins make them notoriously hard to handle in biochemical, biophysical, and structural experiments. Here, we describe the recent advances in computational approaches to design stabilizing mutations for GPCR that take advantage of the structural and sequence conservation properties of the receptors, and employ machine learning on accumulated mutation data for the superfamily,” the investigators wrote.
“The fast and effective computational tools can provide a viable alternative to existing experimental mutation screening and are poised for further improvements with expansion of thermostability datasets for training the machine learning models. The rapidly growing practical applications of computational stability design streamline GPCR structure determination and may contribute to more efficient drug discovery.”
GPCRs are among the best-known human protein families involved in vision, olfaction, immune response, and brain processes, making them an important drug target. For a receptor to serve as a target, the researchers need to understand its structure in great detail, just as a locksmith needs to know the lock’s inner structure to make a key that fits. Studying a receptor that becomes unstable when detached from the cell membrane is a much more challenging task, which is largely facilitated by the computational methods that help to accurately predict the receptor’s soft spots and the changes that will make it more stable.
“The structural studies of GPCRs are of high scientific and applied value, since these proteins are the target for 30–40% of drugs. Our method relies on several approaches, including machine learning, molecular modeling, and bioinformatics, that are tailored specifically to GPCRs. These approaches are complementary, which enables effectively predicting the smallest possible changes that can enhance the receptor’s stability and make it easier to obtain its molecular structure,” explained Petr Popov of MIPT’s Laboratory of Structural Biology of G Protein-Coupled Receptors and the Skoltech Center for Computational and Data-Intensive Science and Engineering.
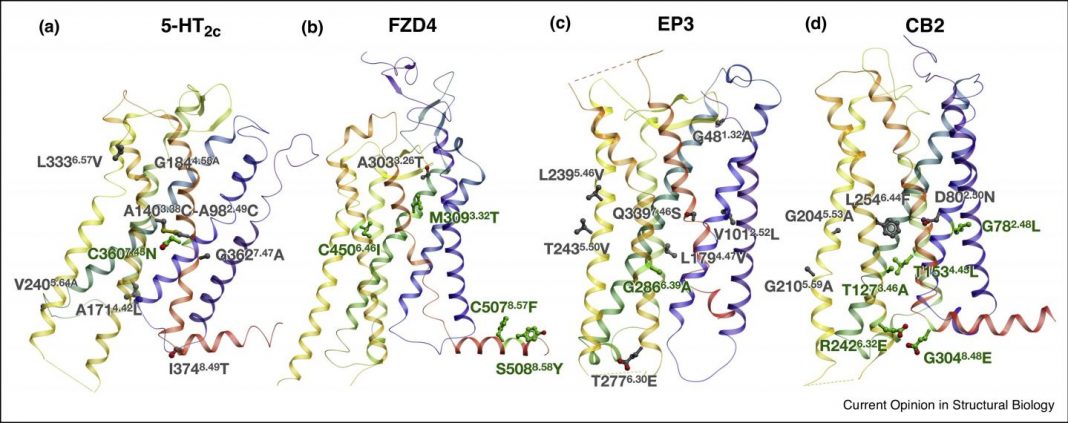
The new method developed at MIPT, Skoltech, and USC allowed researchers to obtain the structures of four important human receptors, including the cannabinoid receptor involved in brain signal transmission and pain perception, and the prostaglandin receptor implicated in inflammatory processes in the human body, added Popov.